UCI Machine Learning Repository Dataset - AIDS Clinical Trials Group Study 175¶
The UCI Machine Learning Repository is a well-known resource for accessing a wide range of datasets used for machine learning research and practice. One such dataset is the AIDS Clinical Trials Group Study dataset, which can be used to build and evaluate predictive models.
In our library, you can easily fetch this dataset using the ucimlrepo package. If you haven't installed it yet, you can do so by running pip install ucimlrepo.
Model Tuner Library Instructions¶
This notebook provides a guide on how to install and use the model_tuner library in a notebook environment like Google Colab.
Model Tuner Description¶
The model_tuner library is designed to streamline the process of hyperparameter tuning and model optimization for machine learning algorithms. It provides an easy-to-use interface for defining, tuning, and evaluating models.
Key Features¶
Automatic Hyperparameter Tuning
The library can automatically tune hyperparameters for a variety of machine learning models using advanced optimization techniques.
Cross-Validation
Integrated cross-validation ensures that the models are evaluated robustly, preventing overfitting.
Documentation¶
For detailed documentation and advanced usage of the model_tuner library, please refer to the model_tuner documentation.
By following these steps, you should be able to install and use the model_tuner library effectively in your notebook environment. If you encounter any issues or have further questions, feel free to reach out for support.
Installation¶
To install the model_tuner library, use the following command:
! pip install model_tuner
Importing the Library¶
After installation, you can import the necessary components from the model_tuner library as shown below:
import model_tuner ## import model_tuner to show version info.
from model_tuner import Model ## Model class from model_tuner lib.
Checking the Version¶
To ensure that the model_tuner library is installed correctly, you can check its version:
print(help(model_tuner))
Binary Classification Via The Breast Cancer Dataset¶
AIDS Clinical Trials Group Study 175 Dataset¶
The AIDS Clinical Trials Group Study 175 Dataset is a healthcare dataset that contains statistical and categorical information about patients who have been diagnosed with AIDS. This dataset, which was initially published in 1996, is often used to predict whether or not a patient will respond to different AIDS treatments.
Key Features of the Dataset¶
- Number of Instances: 2,139
- Number of Features: 23
- Feature Type: Categorical, Integer
- Subject Area: Health and Medicine
- Associated Tasks: Classification, Regression
Dataset Information¶
- Purpose of the Dataset: The dataset was created to examine the performance of two different types of AIDS treatments.
- Funding: The creation of this dataset was funded by the AIDS Clinical Trials Group of the National Institute of Allergy and Infectious Diseases and General Research Center units funded by the National Center for Research Resources.
- Instances Represent: The dataset includes health records of AIDS patients from the US only. Sensitive Data: The dataset includes sensitive information such as ethnicity (race) and gender.
- Data Preprocessing: No preprocessing was performed on the data.
- Missing Values: The dataset does not have missing values.
Example Usage in Machine Learning¶
- Predictive Modeling: The dataset can be used to train models that predict patient outcomes based on demographic and clinical features.
- Treatment Efficacy Analysis: Researchers can use the dataset to compare the effectiveness of different AIDS treatments.
- Health Data Analytics: This dataset is valuable for analyzing trends in the progression and treatment of AIDS among patients in the United States.
Accessing the Dataset¶
To work with the AIDS Clinical Trials Group Study 175 Dataset, you can load it using the ucimlrepo package. If you haven't installed it yet, install it with:
pip install ucimlrepo
! pip install ucimlrepo
Load the dataset, define X, y¶
Once installed, you can quickly load the AIDS Clinical Trials Group Study dataset with a few simple commands:
from ucimlrepo import fetch_ucirepo
## Fetch dataset
aids_clinical_trials_group_study_175 = fetch_ucirepo(id=890)
## Data (as pandas dataframes)
X = aids_clinical_trials_group_study_175.data.features
y = aids_clinical_trials_group_study_175.data.targets
Import Requisite Libraries¶
import pandas as pd
import numpy as np
from xgboost import XGBClassifier
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import StandardScaler
from sklearn.feature_selection import RFE
from sklearn.linear_model import ElasticNet
X.head() ## Inspect the first 5 rows of data
if isinstance(y, pd.DataFrame):
y = y.squeeze()
Check for zero-variance columns and drop accordingly¶
## Check for zero-variance columns and drop them
zero_variance_columns = X.columns[X.var() == 0]
if not zero_variance_columns.empty:
X = X.drop(columns=zero_variance_columns)
Define Hyperparameters for XGBoost¶
xgb_name = "xgb"
xgb = XGBClassifier(
objective="binary:logistic",
random_state=222,
)
xgbearly = True
tuned_parameters_xgb = {
f"{xgb_name}__max_depth": [3, 10, 20, 200, 500],
f"{xgb_name}__learning_rate": [1e-4],
f"{xgb_name}__n_estimators": [1000],
f"{xgb_name}__early_stopping_rounds": [100],
f"{xgb_name}__verbose": [0],
f"{xgb_name}__eval_metric": ["logloss"],
}
xgb_definition = {
"clc": xgb,
"estimator_name": xgb_name,
"tuned_parameters": tuned_parameters_xgb,
"randomized_grid": False,
"n_iter": 5,
"early": xgbearly,
}
Define The Model Object¶
model_type = "xgb"
clc = xgb_definition["clc"]
estimator_name = xgb_definition["estimator_name"]
tuned_parameters = xgb_definition["tuned_parameters"]
n_iter = xgb_definition["n_iter"]
rand_grid = xgb_definition["randomized_grid"]
early_stop = xgb_definition["early"]
kfold = False
calibrate = True
Using Imputation and Scaling in Pipeline Steps for Model Preprocessing¶
The pipeline_steps parameter accepts a list of tuples, where each tuple specifies a transformation step to be applied to the data. For example, the code block below performs imputation followed by standardization on the dataset before training the model.
pipeline_steps=[
("Imputer", SimpleImputer()),
("StandardScaler", StandardScaler()),
]
When Is Imputation and Feature Scaling in pipeline_steps Beneficial?
- Logistic Regression: Highly sensitive to feature scaling and missing data. Preprocessing steps like imputation and standardization improve model performance significantly.
- Linear Models (e.g., Ridge, Lasso): Similar to Logistic Regression, these models require feature scaling for optimal performance.
- SVMs: Sensitive to the scale of the features, requiring preprocessing like standardization.
Models Not Benefiting From Imputation and Scaling in pipeline_steps:
- Tree-Based Models (e.g., XGBoost, Random Forests, Decision Trees): These models are invariant to feature scaling and can handle missing values natively. Passing preprocessing steps like StandardScaler or Imputer may be redundant or even unnecessary.
Why Doesn't XGBoost Require Imputation and Scaling in pipeline_steps?
XGBoost and similar tree-based models work on feature splits rather than feature values directly. This makes them robust to unscaled data and capable of handling missing values using default mechanisms like missing parameter handling in XGBoost. Thus, adding steps like scaling or imputation often does not improve and might complicate the training process.
To this end, it is best to use pipeline_steps strategically for algorithms that rely on numerical properties (e.g., Logistic Regression). For XGBoost, focus on other optimization techniques like hyperparameter tuning and feature engineering instead.
Initialize and Configure the Model¶
model_xgb = Model(
name=f"AIDS_Clinical_{model_type}",
estimator_name=estimator_name,
calibrate=calibrate,
estimator=clc,
model_type="classification",
kfold=kfold,
stratify_y=True,
stratify_cols=False,
grid=tuned_parameters,
randomized_grid=rand_grid,
boost_early=early_stop,
scoring=["roc_auc"],
random_state=222,
n_jobs=2,
)
Tuning f1_beta_tune and optimal_threshold Parameters for Improved Performance on Imbalanced Datasets¶
When working with imbalance datasets, standard metrics like precision and F-score can be misleading, especially for classes with few samples. To address this, we provide the f1_beta_tune and optimal_threshold parameters, which allow for more reliable metric calculation and improved model performance.
f1_beta_tune: Settingf1_beta_tune=Trueenables the model to adjust the F1 beta threshold during parameter tuning. This adjustment lets you control the balance between precision and recall based on the needs of your application. For instance, a higher beta value would prioritize recall, which might be crucial in applications where false negatives are costly. This parameter is particularly valuable for fine-tuning models on imbalanced datasets where a single threshold may not optimize F1 performance across classes.optimal_threshold: By settingoptimal_threshold=Trueinreturn_metrics(), the model will automatically find the threshold that maximizes the F1 score. This dynamic threshold adjustment helps avoid situations where precision or F-score are undefined (set to 0.0) due to a lack of predicted samples for certain classes. This preventsUndefinedMetricWarningand ensures that metrics are calculated more consistently across all classes, even in cases of severe class imbalance.
These parameters enable better handling of imbalanced datasets, providing more reliable metrics and improving overall model interpretability.
Perform Grid Search Parameter Tuning and Retrieve Split Data¶
model_xgb.grid_search_param_tuning(X, y, f1_beta_tune=True)
X_train, y_train = model_xgb.get_train_data(X, y)
X_test, y_test = model_xgb.get_test_data(X, y)
X_valid, y_valid = model_xgb.get_valid_data(X, y)
Fit the Model¶
Model Training with Evaluation Set and Scoring Options¶
In XGBoost, specifying an eval_set (such as validation_data=[X_valid, y_valid]) within the fit method is recommended for monitoring model performance on unseen data during training. This helps in early stopping and in selecting optimal parameters by evaluating model performance metrics at each boosting round.
By default, XGBoost uses ROC AUC as the scoring metric for binary classification, a measure that reflects the trade-off between true positive and false positive rates. However, users can adjust this by setting score="average_precision" if optimizing for precision-recall (particularly useful in imbalanced datasets). This flexibility allows the model to be tailored to specific performance needs based on the application.
model_xgb.fit(
X_train,
y_train,
validation_data=[X_valid, y_valid],
)
Return Metrics (Optional)¶
# ------------------------- VALID AND TEST METRICS -----------------------------
print("Validation Metrics")
class_report_val, cm_val = model_xgb.return_metrics(
X_valid,
y_valid,
optimal_threshold=True,
)
print()
print("Test Metrics")
class_report_test, cm_test = model_xgb.return_metrics(
X_test,
y_test,
optimal_threshold=True,
)
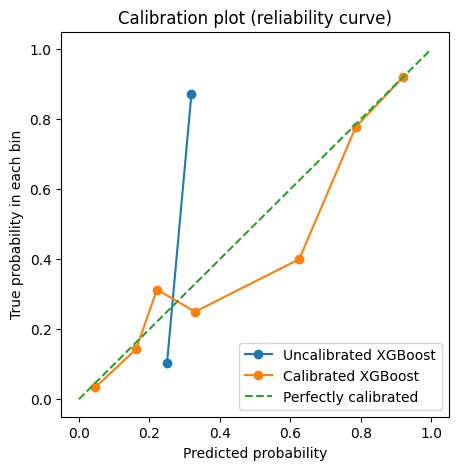
Calibrate the Model¶
import matplotlib.pyplot as plt
from sklearn.calibration import calibration_curve
## Get the predicted probabilities for the validation data from uncalibrated model
y_prob_uncalibrated = model_xgb.predict_proba(X_test)[:, 1]
## Compute the calibration curve for the uncalibrated model
prob_true_uncalibrated, prob_pred_uncalibrated = calibration_curve(
y_test,
y_prob_uncalibrated,
n_bins=10,
)
## Calibrate the model
if model_xgb.calibrate:
model_xgb.calibrateModel(X, y, score="roc_auc")
## Predict on the validation set
y_test_pred = model_xgb.predict_proba(X_test)[:, 1]
## Get the predicted probabilities for the validation data from calibrated model
y_prob_calibrated = model_xgb.predict_proba(X_test)[:, 1]
## Compute the calibration curve for the calibrated model
prob_true_calibrated, prob_pred_calibrated = calibration_curve(
y_test,
y_prob_calibrated,
n_bins=10,
)
## Plot the calibration curves
plt.figure(figsize=(5, 5))
plt.plot(
prob_pred_uncalibrated,
prob_true_uncalibrated,
marker="o",
label="Uncalibrated XGBoost",
)
plt.plot(
prob_pred_calibrated,
prob_true_calibrated,
marker="o",
label="Calibrated XGBoost",
)
plt.plot(
[0, 1],
[0, 1],
linestyle="--",
label="Perfectly calibrated",
)
plt.xlabel("Predicted probability")
plt.ylabel("True probability in each bin")
plt.title("Calibration plot (reliability curve)")
plt.legend()
plt.show()
print(model_xgb.classification_report)
Recursive Feature Elimination (RFE)¶
Now that we've trained the models, we can also refine them by identifying which features contribute most to their performance. One effective method for this is Recursive Feature Elimination (RFE). This technique allows us to systematically remove the least important features, retraining the model at each step to evaluate how performance is affected. By focusing only on the most impactful variables, RFE helps streamline the dataset, reduce noise, and improve both the accuracy and interpretability of the final model.
It works by recursively training a model, ranking the importance of features based on the model’s outputas (such as coefficients in linear models or importance scores in tree-based models), and then removing the least important features one by one. This process continues until a specified number of features remains or the desired performance criteria are met.
The primary advantage of RFE is its ability to streamline datasets, improving model performance and interpretability by focusing on features that contribute the most to the predictive power. However, it can be computationally expensive since it involves repeated model training, and its effectiveness depends on the underlying model’s ability to evaluate feature importance. RFE is commonly used with cross-validation to ensure that the selected features generalize well across datasets, making it a robust choice for model optimization and dimensionality reduction.
As an illustrative example, we will retrain the above model using RFE.
We will begin by appending the feature selection technique to our tuned_parameters dictionary.
xgb_definition["tuned_parameters"][f"feature_selection_rfe__n_features_to_select"] = [
5,
10,
]
We will use ElasticNet because it strikes a balance between two widely used regularization techniques: Lasso (L1) and Ridge (L2). ElasticNet is particularly effective in scenarios where we expect the dataset to have a mix of strongly and weakly correlated features. Lasso alone tends to select only one feature from a group of highly correlated ones, ignoring the others, while Ridge includes all features but may not perform well when some are entirely irrelevant. ElasticNet addresses this limitation by combining both penalties, allowing it to handle multicollinearity more effectively while still performing feature selection.
Additionally, ElasticNet provides flexibility by controlling the ratio between L1 and L2 penalties, enabling fine-tuning to suit the specific needs of our dataset. This makes it a robust choice for datasets with many features, some of which may be irrelevant or redundant, as it can reduce overfitting while retaining a manageable subset of predictors.
rfe_estimator = ElasticNet(alpha=10.0, l1_ratio=0.9)
rfe = RFE(rfe_estimator)
from model_tuner import evaluate_bootstrap_metrics
model_xgb = Model(
name=f"AIDS_Clinical_{model_type}",
estimator_name=estimator_name,
calibrate=calibrate,
estimator=clc,
model_type="classification",
kfold=kfold,
pipeline_steps=[
("rfe", rfe),
],
stratify_y=True,
stratify_cols=False,
grid=tuned_parameters,
randomized_grid=rand_grid,
feature_selection=True,
boost_early=early_stop,
scoring=["roc_auc"],
random_state=222,
n_jobs=2,
)
model_xgb.grid_search_param_tuning(X, y, f1_beta_tune=True)
X_train, y_train = model_xgb.get_train_data(X, y)
X_test, y_test = model_xgb.get_test_data(X, y)
X_valid, y_valid = model_xgb.get_valid_data(X, y)
model_xgb.fit(
X_train,
y_train,
validation_data=[X_valid, y_valid],
)
# ------------------------- VALID AND TEST METRICS -----------------------------
print("Validation Metrics")
model_xgb.return_metrics(
X_valid,
y_valid,
optimal_threshold=True,
)
print()
print("Test Metrics")
model_xgb.return_metrics(
X_test,
y_test,
optimal_threshold=True,
)
print()
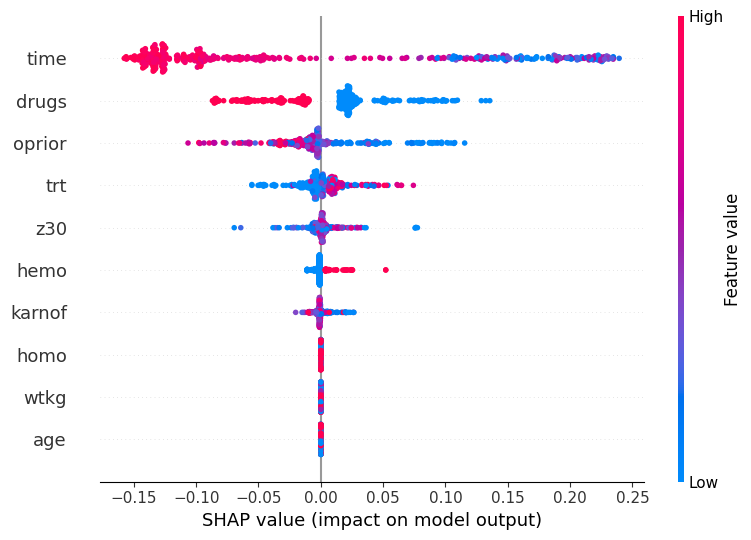
Using SHAP to Interpret Model Predictions with a Pipeline and Feature Selection¶
This example demonstrates how to compute and visualize SHAP (SHapley Additive exPlanations) values for a machine learning model with a pipeline that includes feature selection. SHAP values provide insights into how individual features contribute to the predictions of a model.
Steps
The dataset is transformed through the model's feature selection pipeline to ensure only the selected features are used for SHAP analysis.
The final model (e.g.,
XGBoostclassifier) is retrieved from the custom Model object. This is required because SHAP operates on the underlying model, not the pipeline.SHAP's
TreeExplaineris used to explain the predictions of the XGBoost classifier.SHAP values are calculated for the transformed dataset to quantify the contribution of each feature to the predictions.
A summary plot is generated to visualize the impact of each feature across all data points.
Step 1: Transform the test data using the feature selection pipeline¶
## The pipeline applies preprocessing (e.g., imputation, scaling) and feature
## selection (RFE) to X_test
X_test_transformed = model_xgb.get_feature_selection_pipeline().transform(X_test)
Step 2: Retrieve the trained XGBoost classifier from the pipeline¶
## The last estimator in the pipeline is the XGBoost model
xgb_classifier = model_xgb.estimator[-1]
Step 3: Extract feature names from the training data, and initialize the SHAP explainer for the XGBoost classifier¶
## Import SHAP for model explainability
import shap
## Feature names are required for interpretability in SHAP plots
feature_names = X_train.columns.to_list()
## Initialize the SHAP explainer with the model
explainer = shap.TreeExplainer(xgb_classifier)
Step 4: Compute SHAP values for the transformed test dataset and generate a summary plot of SHAP values¶
## Compute SHAP values for the transformed dataset
shap_values = explainer.shap_values(X_test_transformed)
Step 5: Generate a summary plot of SHAP values¶
## Plot SHAP values
## Summary plot of SHAP values for all features across all data points
shap.summary_plot(shap_values, X_test_transformed, feature_names=feature_names,)
Reference¶
El-Sadr, W., & Abrams, D. (1998). AIDS Clinical Trials Group Study 175. UCI Machine Learning Repository.
https://doi.org/10.24432/C5G896.