Imbalanced Dataset for Classification¶
In machine learning, imbalanced datasets are a frequent challenge, especially in real-world scenarios. These datasets have an unequal distribution of target classes, with one class (e.g., fraudulent transactions, rare diseases, or other low-frequency events) being underrepresented compared to the majority class. Models trained on imbalanced data often struggle to generalize, as they tend to favor the majority class, leading to poor performance on the minority class.
To mitigate these issues, it is crucial to:
- Understand the nature of the imbalance in the dataset.
- Apply appropriate resampling techniques (oversampling, undersampling, or hybrid methods).
- Use metrics beyond accuracy, such as precision, recall, and F1-score, to evaluate model performance fairly.
The model_tuner Library: Simplifying Model Development and Evaluation¶
Before diving into imbalance handling techniques, it's worth mentioning the model_tuner library, a powerful tool designed for streamlining the machine learning model development process.
Model Tuner Library Instructions¶
This notebook provides a guide on how to install and use the model_tuner library in a notebook environment like Google Colab.
Model Tuner Description¶
The model_tuner library is designed to streamline the process of hyperparameter tuning and model optimization for machine learning algorithms. It provides an easy-to-use interface for defining, tuning, and evaluating models.
Key Features¶
Automatic Hyperparameter Tuning
The library can automatically tune hyperparameters for a variety of machine learning models using advanced optimization techniques.
Cross-Validation
Integrated cross-validation ensures that the models are evaluated robustly, preventing overfitting.
Documentation¶
For detailed documentation and advanced usage of the model_tuner library, please refer to the model_tuner documentation.
By following these steps, you should be able to install and use the model_tuner library effectively in your notebook environment. If you encounter any issues or have further questions, feel free to reach out for support.
Installation¶
To install the model_tuner library, use the following command:
! pip install model_tuner
Importing the Library¶
After installation, you can import the necessary components from the model_tuner library as shown below:
import model_tuner # import model_tuner to show version info.
from model_tuner import Model # Model class from model_tuner lib.
Checking the Version¶
To ensure that the model_tuner library is installed correctly, you can check its version:
print(help(model_tuner))
Overview¶
In machine learning, imbalanced datasets are common in real-world scenarios, where the distribution of classes in a dataset is not uniform. For instance, in fraud detection, disease diagnosis, or rare event prediction, one class (e.g., fraudulent transactions or diseased cases) often occurs significantly less frequently than the other. Training machine learning models on such datasets can lead to biases towards the majority class, making it essential to create, understand, and handle imbalanced datasets effectively during model development.
Import Requisite Libraries¶
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import make_classification
from xgboost import XGBClassifier
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import StandardScaler
from imblearn.over_sampling import SMOTE, ADASYN
from imblearn.under_sampling import RandomUnderSampler
Generating an Imbalanced Dataset¶
Demonstrated below are the steps to generate an imbalanced dataset using make_classification from the sklearn.datasets module. The following parameters are specified:
n_samples=1000: The dataset contains 1,000 samples.
n_features=20: Each sample has 20 features.
n_informative=2: Two features are informative for predicting the target.
n_redundant=2: Two features are linear combinations of the informative features.
weights=[0.9, 0.1]: The target class distribution is 90% for the majority class and 10% for the minority class, creating an imbalance.
flip_y=0: No label noise is added to the target variable.
random_state=42: Ensures reproducibility by using a fixed random seed.
# Create an imbalanced dataset using make_classification
X, y = make_classification(
n_samples=1000, # Total number of samples
n_features=20, # Total number of features
n_informative=2, # Number of informative features
n_redundant=2, # Number of redundant features
n_clusters_per_class=1,
weights=[0.9, 0.1], # Proportion of classes, creating imbalance
flip_y=0, # No label noise
random_state=42,
)
# Convert to a pandas DataFrame for better visualization
data = pd.DataFrame(X, columns=[f'feature_{i}' for i in range(1, 21)])
data['target'] = y
X = data[[col for col in data.columns if "target" not in col]]
y = pd.Series(data["target"])
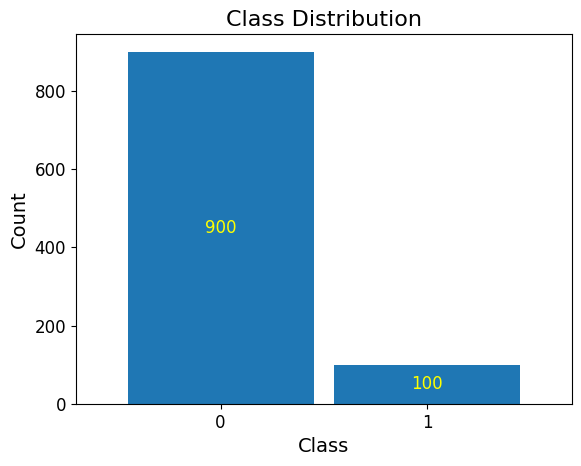
Imbalanced Dataset¶
Below, you will see that the dataset we have generated is severely imbalanced with 900 observations allocated to the majority class (0) and 100 observations to the minority class (1).
## Create a bar plot
value_counts = pd.Series(y).value_counts()
ax = value_counts.plot(
kind="bar",
rot=0,
width=0.9,
)
## Add labels inside the bars
for index, count in enumerate(value_counts):
plt.text(
index,
count / 2,
str(count),
ha="center",
va="center",
color="yellow",
fontsize=12,
)
## Customize labels and title
plt.xlabel("Class", fontsize=14)
plt.ylabel("Count", fontsize=14)
plt.title("Class Distribution", fontsize=16)
## Adjust tick label size
ax.tick_params(axis='x', labelsize=12)
ax.tick_params(axis='y', labelsize=12)
## Show the plot
plt.show()
Define Hyperparameters for XGBoost¶
Below, we will use an XGBoost classifier with the following hyperparameters:
xgb_name = "xgb"
xgb = XGBClassifier(
# objective="binary:logistic",
random_state=222,
)
xgbearly = True
tuned_parameters_xgb = {
f"{xgb_name}__max_depth": [3, 10, 20, 200, 500],
f"{xgb_name}__learning_rate": [1e-4],
f"{xgb_name}__n_estimators": [1000],
f"{xgb_name}__early_stopping_rounds": [100],
f"{xgb_name}__verbose": [0],
f"{xgb_name}__eval_metric": ["logloss"],
}
xgb_definition = {
"clc": xgb,
"estimator_name": xgb_name,
"tuned_parameters": tuned_parameters_xgb,
"randomized_grid": False,
"n_iter": 5,
"early": xgbearly,
}
Define The Model Object¶
model_type = "xgb"
clc = xgb_definition["clc"]
estimator_name = xgb_definition["estimator_name"]
tuned_parameters = xgb_definition["tuned_parameters"]
n_iter = xgb_definition["n_iter"]
rand_grid = xgb_definition["randomized_grid"]
early_stop = xgb_definition["early"]
kfold = False
calibrate = True
Addressing Class Imbalance in Machine Learning¶
Class imbalance occurs when one class significantly outweighs another in the dataset, leading to biased models that perform well on the majority class but poorly on the minority class. Techniques like SMOTE and others aim to address this issue by improving the representation of the minority class, ensuring balanced learning and better generalization.
Techniques to Address Class Imbalance¶
Resampling Techniques
SMOTE (Synthetic Minority Oversampling Technique): SMOTE generates synthetic samples for the minority class by interpolating between existing minority class data points and their nearest neighbors. This helps create a more balanced class distribution without merely duplicating data, thus avoiding overfitting.
Oversampling: Randomly duplicates examples from the minority class to balance the dataset. While simple, it risks overfitting to the duplicated examples.
Undersampling: Reduces the majority class by randomly removing samples. While effective, it can lead to loss of important information.
Purpose of Using These Techniques¶
The goal of using these techniques is to improve model performance on imbalanced datasets, specifically by:
- Ensuring the model captures meaningful patterns in the minority class.
- Reducing bias toward the majority class, which often dominates predictions in imbalanced datasets.
- Improving metrics like recall, F1-score, and AUC-ROC for the minority class, which are critical in applications like fraud detection, healthcare, and rare event prediction.
Synthetic Minority Oversampling Technique (SMOTE)¶
SMOTE (Synthetic Minority Oversampling Technique) is a method used to address class imbalance in datasets. It generates synthetic samples for the minority class by interpolating between existing minority samples and their nearest neighbors, effectively increasing the size of the minority class without duplicating data. This helps models better learn patterns from the minority class, improving classification performance on imbalanced datasets.
Initialize and Configure the Model¶
xgb_smote = Model(
name=f"Make_Classification_{model_type}",
estimator_name=estimator_name,
calibrate=calibrate,
model_type="classification",
estimator=clc,
kfold=kfold,
stratify_y=True,
stratify_cols=False,
grid=tuned_parameters,
randomized_grid=rand_grid,
boost_early=early_stop,
scoring=["roc_auc"],
random_state=222,
n_jobs=2,
imbalance_sampler=SMOTE(),
)
Perform Grid Search Parameter Tuning and Retrieve Split Data¶
xgb_smote.grid_search_param_tuning(
X,
y,
f1_beta_tune=True,
)
X_train, y_train = xgb_smote.get_train_data(X, y)
X_test, y_test = xgb_smote.get_test_data(X, y)
X_valid, y_valid = xgb_smote.get_valid_data(X, y)
SMOTE: Distribution of y values after resampling¶
Notice that the target has been redistributed after SMOTE to 540 observations for the minority class and 540 observations for the majority class.
Fit The Model¶
xgb_smote.fit(
X_train,
y_train,
validation_data=[X_valid, y_valid],
)
Return Metrics (Optional)¶
# ------------------------- VALID AND TEST METRICS -----------------------------
print("Validation Metrics")
class_report_val, cm_val = xgb_smote.return_metrics(
X_valid,
y_valid,
optimal_threshold=True,
)
print()
print("Test Metrics")
class_report_test, cm_test = xgb_smote.return_metrics(
X_test,
y_test,
optimal_threshold=True,
)
Adaptive Synthetic Oversampling (ADASYN)¶
ADASYN (Adaptive Synthetic Sampling) is a variation of SMOTE that also generates synthetic samples for the minority class but focuses on harder-to-learn instances. It adaptively determines the number of synthetic samples to generate for each minority instance based on the degree of difficulty (i.e., the density of the majority class in its neighborhood).
ADASYN aims to create a balanced dataset while improving the model's ability to distinguish between classes, especially in complex or overlapping regions. This technique is particularly useful when dealing with highly imbalanced data with challenging decision boundaries.
xgb_adasyn = Model(
name=f"Make_Classification_{model_type}",
estimator_name=estimator_name,
calibrate=calibrate,
model_type="classification",
estimator=clc,
kfold=kfold,
stratify_y=True,
stratify_cols=False,
grid=tuned_parameters,
randomized_grid=rand_grid,
boost_early=early_stop,
scoring=["roc_auc"],
random_state=222,
n_jobs=2,
imbalance_sampler=ADASYN(),
)
Perform Grid Search Parameter Tuning and Retrieve Split Data¶
xgb_adasyn.grid_search_param_tuning(
X,
y,
f1_beta_tune=True,
),
X_train, y_train = xgb_adasyn.get_train_data(X, y)
X_test, y_test = xgb_adasyn.get_test_data(X, y)
X_valid, y_valid = xgb_adasyn.get_valid_data(X, y)
ADASYN: Distribution of y values after resampling¶
Notice that the target has been redistributed after ADASYN to 540 observations for the minority class and 540 observations for the majority class.
Fit The Model¶
xgb_adasyn.fit(
X_train,
y_train,
validation_data=[X_valid, y_valid],
)
Return Metrics (Optional)¶
# ------------------------- VALID AND TEST METRICS -----------------------------
print("Validation Metrics")
class_report_val, cm_val = xgb_adasyn.return_metrics(
X_valid,
y_valid,
optimal_threshold=True,
)
print()
print("Test Metrics")
class_report_test, cm_test = xgb_adasyn.return_metrics(
X_test,
y_test,
optimal_threshold=True,
)
Random Undersampler¶
Random Undersampling is a technique used to address class imbalance by reducing the number of samples in the majority class. It randomly removes instances from the majority class until the class distribution is balanced or reaches a desired ratio.
Random undersampling is used to balance datasets, enabling models to focus equally on both classes and reducing bias toward the majority class. It is often combined with oversampling techniques (e.g., SMOTE) to mitigate the risk of losing important data.
xgb_undersampled = Model(
name=f"Make_Classification_{model_type}",
estimator_name=estimator_name,
calibrate=calibrate,
model_type="classification",
estimator=clc,
kfold=kfold,
stratify_y=True,
stratify_cols=False,
grid=tuned_parameters,
randomized_grid=rand_grid,
boost_early=early_stop,
scoring=["roc_auc"],
random_state=222,
n_jobs=2,
imbalance_sampler=RandomUnderSampler(),
)
Perform Grid Search Parameter Tuning and Retrieve Split Data¶
xgb_undersampled.grid_search_param_tuning(
X,
y,
f1_beta_tune=True,
)
X_train, y_train = xgb_undersampled.get_train_data(X, y)
X_test, y_test = xgb_undersampled.get_test_data(X, y)
X_valid, y_valid = xgb_undersampled.get_valid_data(X, y)
Random Undersampler: Distribution of y values after resampling¶
Notice that the target has been redistributed after Random Undersampler to 60 observations for the minority class and 60 observations for the majority class.
Fit The Model¶
xgb_undersampled.fit(
X_train,
y_train,
validation_data=[X_valid, y_valid],
)
Return Metrics (Optional)¶
# ------------------------- VALID AND TEST METRICS -----------------------------
print("Validation Metrics")
class_report_val, cm_val = xgb_undersampled.return_metrics(
X_valid,
y_valid,
optimal_threshold=True,
)
print()
print("Test Metrics")
class_report_test, cm_test = xgb_undersampled.return_metrics(
X_test,
y_test,
optimal_threshold=True,
)