iPython Notebooks
Binary Classification Examples
Regression Example
Google Colab Notebook
HTML File
Note
This class is designed to be flexible and can be extended to include additional functionalities or custom metrics.
It is essential to properly configure the parameters during initialization to suit the specific requirements of your machine learning task.
Ensure that all dependencies are installed and properly imported before using the
Modelclass from themodel_tunerlibrary.
Input Parameters
- class Model(name, estimator_name, estimator, model_type, calibrate=False, kfold=False, imbalance_sampler=None, train_size=0.6, validation_size=0.2, test_size=0.2, stratify_y=False, stratify_cols=None, grid=None, scoring=['roc_auc'], n_splits=10, random_state=3, n_jobs=1, display=True, randomized_grid=False, n_iter=100, pipeline_steps=[], boost_early=False, feature_selection=False, class_labels=None, multi_label=False, calibration_method='sigmoid', custom_scorer=[], bayesian=False)
A class for building, tuning, and evaluating machine learning models, supporting both classification and regression tasks, as well as multi-label classification.
- Parameters:
name (str) – A unique name for the model, helpful for tracking outputs and logs.
estimator_name (str) – Prefix for the estimator in the pipeline, used for setting parameters in tuning (e.g., estimator_name +
__param_name).estimator (object) – The machine learning model to be trained and tuned.
model_type (str) – Specifies the type of model, must be either
classificationorregression.calibrate (bool, optional) – Whether to calibrate the model’s probability estimates. Default is
False.kfold (bool, optional) – Whether to perform k-fold cross-validation. Default is
False.imbalance_sampler (object, optional) – An imbalanced data sampler from the imblearn library, e.g.,
RandomUnderSamplerorRandomOverSampler.train_size (float, optional) – Proportion of the data to be used for training. Default is
0.6.validation_size (float, optional) – Proportion of the data to be used for validation. Default is
0.2.test_size (float, optional) – Proportion of the data to be used for testing. Default is
0.2.stratify_y (bool, optional) – Whether to stratify by the target variable during data splitting. Default is
False.stratify_cols (str, list, or pandas.DataFrame, optional) – Columns to use for stratification during data splitting. Can be a single column name (as a string), a list of column names (as strings), or a DataFrame containing the columns for stratification. Default is
None.grid (list of dict) – Hyperparameter grid for model tuning, supporting both regular and Bayesian search.
scoring (list of str) – List of scoring metrics for evaluation, e.g.,
["roc_auc", "accuracy"].n_splits (int, optional) – Number of splits for k-fold cross-validation. Default is
10.random_state (int, optional) – Seed for random number generation to ensure reproducibility. Default is
3.n_jobs (int, optional) – Number of parallel jobs to run for model fitting. Default is
1.display (bool, optional) – Whether to print messages during the tuning and training process. Default is
True.randomized_grid (bool, optional) – Whether to use randomized grid search. Default is
False.n_iter (int, optional) – Number of iterations for randomized grid search. Default is
100.pipeline_steps (list, optional) – List of steps for the pipeline, e.g., preprocessing and feature selection steps. Default is
[].boost_early (bool, optional) – Whether to enable early stopping for boosting algorithms like XGBoost. Default is
False.feature_selection (bool, optional) – Whether to enable feature selection. Default is
False.class_labels (list, optional) – List of labels for multi-class classification. Default is
None.multi_label (bool, optional) – Whether the task is a multi-label classification problem. Default is
False.calibration_method (str, optional) – Method for calibration; options include
sigmoidandisotonic. Default issigmoid.custom_scorer (dict, optional) – Dictionary of custom scoring functions, allowing additional metrics to be evaluated. Default is
[].bayesian (bool, optional) – Whether to perform Bayesian hyperparameter tuning using
BayesSearchCV. Default isFalse.
- Raises:
ImportError – If the
bootstrappermodule is not found or not installed.ValueError – Raised for various issues, such as: - Invalid
model_typevalue. Themodel_typemust be explicitly specified as eitherclassificationorregression. - Invalid hyperparameter configurations or mismatchedXandyshapes.AttributeError – Raised if an expected pipeline step is missing, or if
self.estimatoris improperly initialized.TypeError – Raised when an incorrect parameter type is provided, such as passing
Noneinstead of a valid object.IndexError – Raised for indexing issues, particularly in confusion matrix formatting functions.
KeyError – Raised when accessing dictionary keys that are not available, such as missing scores in
self.best_params_per_score.RuntimeError – Raised for unexpected issues during model fitting or transformations that do not fit into the other exception categories.
Key Methods and Functionalities
__init__(...)
Initializes the model tuner with configurations, including estimator, cross-validation settings, scoring metrics, pipeline steps, feature selection, imbalance sampler, Bayesian search, and model calibration options.
reset_estimator()
- reset_estimator()
Resets the estimator and pipeline configuration.
Description:
This function reinitializes the
estimatorattribute of the class based on the current pipeline configuration.If
pipeline_stepsare defined, it creates a new pipeline usingself.PipelineClassand a deep copy of the steps.If
pipeline_stepsare not defined, it resets theestimatorto a single-step pipeline containing the original estimator.
Behavior:
If
self.pipeline_stepsis not empty:Creates a pipeline using the defined steps.
If
self.pipeline_stepsis empty:Resets the
estimatorto a single-step pipeline with the original estimator.
Attributes Used:
self.pipeline_steps: The steps of the pipeline (if defined).self.PipelineClass: The class used to construct pipelines.self.estimator_name: The name of the primary estimator step.self.original_estimator: The original estimator to be reset.
Output:
The function updates the
self.estimatorattribute and does not return a value.
Notes:
This function is intended for internal use as a helper function to manage pipeline and estimator states.
Ensures that the pipeline or estimator is always in a valid state after modifications or resets.
process_imbalance_sampler()
- process_imbalance_sampler(X_train, y_train)
Processes the imbalance sampler, applying it to resample the training data.
- Parameters:
X_train (
pandas.DataFrameor array-like) – Training features to be resampled.y_train (
pandas.Seriesor array-like) – Training target labels to be resampled.
- Raises:
KeyError – Raised if the
resamplerstep is missing in the pipeline.ValueError – Raised if
X_trainory_trainare incompatible with the pipeline or resampler.
Output:
Prints the class distribution of
y_trainafter resampling.Does not modify the original
X_trainory_train.
Description:
This function applies an imbalance sampler to resample the training data, ensuring the target distribution is balanced.
If preprocessing steps are defined in the pipeline, they are applied to the training features before resampling.
Prints the distribution of
y_trainafter resampling to provide visibility into the balance of classes.
Behavior:
If preprocessing steps exist (
self.pipeline_steps):Applies preprocessing to
X_trainusing the preprocessing pipeline obtained fromget_preprocessing_pipeline().
Clones the
resamplerstep from the pipeline to ensure independent operation.Resamples the training data using the cloned resampler, modifying the distribution of
y_train.
Attributes Used:
self.pipeline_steps: Indicates whether preprocessing steps are defined.self.get_preprocessing_pipeline(): Retrieves the preprocessing pipeline (if available).self.estimator.named_steps["resampler"]: The resampler to apply for balancing the target classes.
Note
The function assumes that the pipeline includes a valid
resamplerstep undernamed_steps.Ensures compatibility with
pandas.DataFrameand array-like structures fory_train.Prints the class distribution of
y_trainafter resampling for user awareness.
calibrateModel()
- calibrateModel(X, y, score=None)
Calibrates the model with cross-validation support and configurable calibration methods, improving probability estimates.
- Parameters:
- Raises:
ValueError – Raised if incompatible parameters (e.g., invalid scoring metric) are passed.
KeyError – Raised if required attributes or parameters are missing.
Description:
Supports model calibration with both k-fold cross-validation and a pre-split train-validation-test workflow.
Uses
CalibratedClassifierCVfor calibration with methods such assigmoidorisotonic(defined byself.calibration_method).Handles cases where imbalance sampling or early stopping is applied during training.
Provides additional support for CPU/GPU device management if applicable.
Behavior:
With K-Fold Cross-Validation:
Resets the estimator to avoid conflicts with pre-calibrated models.
Calibrates the model using k-fold splits with the configured calibration method.
Optionally evaluates calibration using the provided scoring metric(s).
Generates and prints confusion matrices for each fold (if applicable).
Without K-Fold Cross-Validation:
Performs a train-validation-test split using
train_val_test_split.Resets the estimator and applies preprocessing or imbalance sampling if configured.
Fits the model on training data, with or without early stopping.
Calibrates the pre-trained model on the test set and evaluates calibration results.
Attributes Used:
self.kfold: Indicates whether k-fold cross-validation is enabled.self.calibrate: Determines whether calibration is applied.self.calibration_method: Specifies the calibration method (e.g.,sigmoidorisotonic).self.best_params_per_score: Stores the best parameters for each scoring metric.self.n_splits: Number of splits for cross-validation.self.stratify_y,self.stratify_cols: Used for stratified train-validation-test splitting.self.imbalance_sampler: Indicates if an imbalance sampler is applied.self.boost_early: Enables early stopping during training.
Output:
Modifies the class attribute
self.estimatorto include the calibrated model.Generates calibration reports and scoring metrics if applicable.
Prints performance metrics (e.g., scores and confusion matrices) for the calibrated model.
Note
When
scoreis provided, the function evaluates calibration using the specified metric(s).Requires the estimator to be compatible with
CalibratedClassifierCV.Handles both balanced and imbalanced datasets with preprocessing support.
Get train, val, test data
Description:
These functions return subsets of the dataset (features and labels) based on predefined indices stored in the class attributes:
self.X_train_indexandself.y_train_indexfor training data.self.X_valid_indexandself.y_valid_indexfor validation data.self.X_test_indexandself.y_test_indexfor test data.
Designed to work with
pandas.DataFrameandpandas.Seriesobjects.
get_train_data()
- get_train_data(X, y)
Retrieves the training data based on specified indices.
- Parameters:
X (
pandas.DataFrame) – Full dataset containing features.y (
pandas.Series) – Full dataset containing target labels.
- Returns:
A tuple containing the training features and labels.
- Return type:
tuple of (
pandas.DataFrame,pandas.Series)
get_valid_data()
- get_valid_data(X, y)
Retrieves the validation data based on specified indices.
- Parameters:
X (
pandas.DataFrame) – Full dataset containing features.y (
pandas.Series) – Full dataset containing target labels.
- Returns:
A tuple containing the validation features and labels.
- Return type:
tuple of (
pandas.DataFrame,pandas.Series)
get_test_data()
- get_test_data(X, y)
Retrieves the test data based on specified indices.
- Parameters:
X (
pandas.DataFrame) – Full dataset containing features.y (
pandas.Series) – Full dataset containing target labels.
- Returns:
A tuple containing the test features and labels.
- Return type:
tuple of (
pandas.DataFrame,pandas.Series)
Note
These methods assume that the indices (e.g.,
self.X_train_index) are defined and valid.The methods return subsets of the provided
Xandydata by indexing the rows based on the stored indices.Useful for workflows where train, validation, and test splits are dynamically managed or predefined.
calibrate_report()
- calibrate_report(X, y, score=None)
Generates a calibration report, including a confusion matrix and classification report.
- Parameters:
X (
pandas.DataFrameor array-like) – Features dataset for validation.y (
pandas.Seriesor array-like) – True labels for the validation dataset.score (str, optional) – Optional scoring metric name to include in the report. Default is
None.
- Raises:
ValueError – Raised if the provided
Xoryare incompatible with the model or metrics.
Description:
This method evaluates the performance of a calibrated model on the validation dataset.
Generates and prints: - A confusion matrix, with support for multi-label classification if applicable. - A classification report summarizing precision, recall, and F1-score for each class.
Behavior:
Calls the
predictmethod to obtain predictions for the validation dataset.Uses
confusion_matrixormultilabel_confusion_matrixbased on the value ofself.multi_labelto compute the confusion matrix.Prints a labeled confusion matrix using the
_confusion_matrix_print()function.Generates a classification report using
classification_reportfromsklearn.metricsand assigns it to theself.classification_reportattribute.
Output:
Prints the following to the console:
The confusion matrix with labels.
The classification report.
A separator line for readability.
Updates the attribute
self.classification_reportwith the generated classification report.
Note
If the model is multi-label, a confusion matrix is generated for each label.
The optional
scoreparameter can be used to specify and display a scoring metric in the report heading.Designed to work with models that support binary, multi-class, or multi-label predictions.
fit()
- fit(X, y, validation_data=None, score=None)
Fits the model to training data and, if applicable, tunes thresholds and performs early stopping. Allows feature selection and processing steps as part of the pipeline.
- Parameters:
X (
pandas.DataFrameor array-like) – Training features.y (
pandas.Seriesor array-like) – Training target labels.validation_data (tuple of (
pandas.DataFrame,pandas.Series), optional) – Tuple containing validation features and labels. Required for early stopping. Default isNone.score (str, optional) – Optional scoring metric to guide the fitting process. Default is
None.
- Raises:
ValueError – Raised if invalid scoring metrics or parameters are provided.
Description:
This method trains the model with support for both k-fold cross-validation and single train-validation-test workflows.
If feature selection or preprocessing steps are configured, they are applied before fitting.
For certain estimators, early stopping is supported when validation data is provided.
The method dynamically sets model parameters based on tuning results for the specified or default scoring metric.
Behavior:
With K-Fold Cross-Validation:
Resets the estimator and fits the model using k-fold splits.
If a scoring metric is provided, applies it to guide the cross-validation.
Stores cross-validation results in the self.xval_output attribute.
Without K-Fold Cross-Validation:
Resets the estimator and applies feature selection or preprocessing if configured.
Fits the model on training data. If early stopping is enabled, uses validation data to monitor performance and stop training early.
Attributes Used:
self.kfold: Indicates whether k-fold cross-validation is enabled.self.best_params_per_score: Stores tuned parameters for different scoring metrics.self.feature_selection,self.pipeline_steps: Flags for feature selection and preprocessing steps.self.imbalance_sampler: Specifies whether imbalance sampling is applied.self.boost_early: Enables early stopping during training.self.estimator_name: Name of the estimator in the pipeline.
Output:
Updates the class attribute
self.estimatorwith the fitted model.For k-fold cross-validation, stores results in
self.xval_output.
Note
Early stopping requires both validation features and labels.
Feature selection and preprocessing steps are dynamically applied based on the pipeline configuration.
When a custom scoring metric is specified, it must match one of the predefined or user-defined metrics.
return_metrics()
- return_metrics(X, y, optimal_threshold=False, model_metrics=False, print_threshold=False, return_dict=False, print_per_fold=False)
A flexible function to evaluate model performance by generating classification or regression metrics. It provides options to print confusion matrices, classification reports, and regression metrics, and supports optimal threshold display and dictionary outputs.
- Parameters:
X (
pandas.DataFrameor array-like) – The feature matrix for evaluation.y (
pandas.Seriesor array-like) – The target vector for evaluation.optimal_threshold (bool, optional) – Whether to use the optimal threshold for predictions (classification only). Default is
False.model_metrics (bool, optional) – Whether to calculate and print detailed model metrics using
report_model_metrics(). Default isFalse.print_threshold (bool, optional) – Whether to print the optimal threshold used for predictions (classification only). Default is
False.return_dict (bool, optional) – Whether to return the metrics as a dictionary instead of printing them. Default is
False.print_per_fold (bool, optional) – For cross-validation, whether to print metrics for each fold. Default is
False.
- Returns:
A dictionary containing metrics if
return_dict=True; otherwise, the metrics are printed.- Return type:
dict or None
Description:
The
return_metrics()function is designed to be highly adaptable, allowing users to:Print Classification Metrics: Displays a confusion matrix and the accompanying classification report when evaluating a classification model.
Print Regression Metrics: Outputs standard regression metrics (e.g., R², Mean Absolute Error) when evaluating a regression model.
Report Detailed Model Metrics: By setting
model_metrics=True, the function invokesreport_model_metrics()to provide detailed insights into metrics like precision, recall, and AUC-ROC.Display the Optimal Threshold: Setting
print_threshold=Truedisplays the threshold value used for classification predictions, particularly when an optimal threshold has been tuned.Return Results as a Dictionary: If
return_dict=True, the metrics are returned in a structured dictionary, allowing users to programmatically access the results. This is especially useful for further analysis or logging.
Behavior:
Classification Models:
Generates and prints a confusion matrix.
Prints a detailed classification report, including precision, recall, F1-score, and accuracy.
Optionally prints additional model metrics and the optimal threshold.
Regression Models:
Outputs standard regression metrics such as R², Mean Absolute Error, and Root Mean Squared Error.
Cross-Validation:
For k-fold validation, the function aggregates metrics across folds and prints the averaged results. If
print_per_fold=True, metrics for each fold are also printed in addition to the averaged results.
Output:
If
return_dict=True, returns:Classification Models:
A dictionary with:
Classification Report: The classification report as a string.Confusion Matrix: The confusion matrix as an array.Best Features: (Optional) List of the top features if feature selection is enabled.
Regression Models:
A dictionary with:
Regression Report: A dictionary of regression metrics.Best Features: (Optional) List of the top features if feature selection is enabled.
If
return_dict=False, prints the metrics directly to the console.
Examples:
## Example usage for validation metrics: print("Validation Metrics") model.return_metrics( X=X_valid, y=y_valid, optimal_threshold=True, print_threshold=True, model_metrics=True, ) print() ## Example usage for test metrics: print("Test Metrics") model.return_metrics( X=X_test, y=y_test, optimal_threshold=True, print_threshold=True, model_metrics=True, ) print()
Note
This function is suitable for both classification and regression models.
Supports cross-validation workflows by calculating metrics across multiple folds.
Enables users to programmatically access metrics via the dictionary output for custom analysis.
predict()
- predict(X, y=None, optimal_threshold=False)
Makes predictions and predicts probabilities, allowing threshold tuning.
- Parameters:
X (
pandas.DataFrameor array-like) – The feature matrix for prediction.y (
pandas.Seriesor array-like, optional) – The true target labels, required only for k-fold predictions. Default isNone.optimal_threshold (bool, optional) – Whether to use an optimal classification threshold for predictions. Default is
False.
- Returns:
Predicted class labels or predictions adjusted by the optimal threshold.
- Return type:
numpy.ndarrayor array-like- Raises:
ValueError – Raised if invalid inputs or configurations are provided.
Description:
Predicts target values for the input data.
Supports both regression and classification tasks, with specific behavior for each:
For regression: Direct predictions are returned, ignoring thresholds.
For classification: Predictions are adjusted using an optimal threshold when enabled.
If k-fold cross-validation is active, performs predictions for each fold using
cross_val_predict.
Behavior:
With K-Fold Cross-Validation:
Returns predictions based on cross-validated folds.
Without K-Fold Cross-Validation:
Uses the trained model’s
predict()method.Applies the optimal threshold to adjust classification predictions, if specified.
Related Methods:
predict_proba(X, y=None):Predicts probabilities for classification tasks.
Supports k-fold cross-validation using
cross_val_predictwith themethod="predict_proba"option.
Note
Optimal thresholding is useful for fine-tuning classification performance metrics such as F1-score or precision-recall balance.
For classification, the threshold can be tuned for specific scoring metrics (e.g., ROC-AUC).
Works seamlessly with pipelines or directly with the underlying model.
grid_search_param_tuning()
- grid_search_param_tuning(X, y, f1_beta_tune=False, betas=[1, 2])
Performs grid or Bayesian search parameter tuning, optionally tuning F-beta score thresholds for classification.
- Parameters:
X (
pandas.DataFrameor array-like) – The feature matrix for training and validation.y (
pandas.Seriesor array-like) – The target vector corresponding toX.f1_beta_tune (bool, optional) – Whether to tune F-beta score thresholds during parameter search. Default is
False.betas (list of int, optional) – List of beta values to use for F-beta score tuning. Default is
[1, 2].
- Raises:
ValueError – Raised if the provided data or configurations are incompatible with parameter tuning.
KeyError – Raised if required scoring metrics are missing.
Description:
This method tunes hyperparameters for a model using grid search or Bayesian optimization.
Supports tuning F-beta thresholds for classification tasks.
Can handle both k-fold cross-validation and single train-validation-test workflows.
Behavior:
With K-Fold Cross-Validation:
Splits data into k folds using
kfold_splitand performs parameter tuning.Optionally tunes thresholds for F-beta scores on validation splits.
Without K-Fold Cross-Validation:
Performs a train-validation-test split using
train_val_test_split.Applies preprocessing, feature selection, and imbalance sampling if configured.
Tunes parameters and thresholds based on validation scores.
Attributes Used:
self.kfold: Indicates whether k-fold cross-validation is enabled.self.scoring: List of scoring metrics used for evaluation.self.best_params_per_score: Stores the best parameter set for each scoring metric.self.grid: Parameter grid for tuning.self.calibrate: Specifies whether the model calibration is enabled.self.imbalance_sampler: Indicates if imbalance sampling is applied.self.feature_selection: Specifies whether feature selection is applied.self.pipeline_steps: Configuration for preprocessing steps.self.boost_early: Enables early stopping during model training.self.threshold: Stores tuned thresholds for F-beta score optimization.
Output:
Updates the class attribute
self.best_params_per_scorewith the best parameters and scores for each metric.Optionally updates
self.thresholdwith tuned F-beta thresholds.Prints best parameters and scores if
self.displayis enabled.
Note
Threshold tuning requires classification tasks and is not applicable for regression.
Early stopping is supported if
self.boost_earlyis enabled and validation data is provided.Works seamlessly with pipelines for preprocessing and feature selection.
print_selected_best_features()
- print_selected_best_features(X)
Prints and returns the selected top K best features based on the feature selection step.
- Parameters:
X (
pandas.DataFrameor array-like) – The feature matrix used during the feature selection process.- Returns:
A list of the selected features or column indices.
- Return type:
- Raises:
AttributeError – Raised if the feature selection pipeline is not properly configured or trained.
Description:
This method retrieves the top K features selected by the feature selection pipeline.
Prints the names or column indices of the selected features to the console.
Returns the selected features as a list.
Behavior:
For DataFrames:
Prints the names of the selected feature columns.
Returns a list of column names corresponding to the selected features.
For Array-like Data:
Prints the indices of the selected feature columns.
Returns a list of column indices.
Attributes Used:
self.get_feature_selection_pipeline(): Retrieves the feature selection pipeline used for selecting features.
Output:
Prints the selected features or indices to the console.
Returns the selected features as a list.
Note
Assumes that a feature selection pipeline has been configured and trained prior to calling this method.
Designed to work with both
pandas.DataFrameand array-like structures for feature matrices.
tune_threshold_Fbeta()
- tune_threshold_Fbeta(score, y_valid, betas, y_valid_proba, kfold=False)
Tunes classification threshold for optimal F-beta score, balancing precision and recall across various thresholds.
- Parameters:
score (str) – A label or name for the score used to store the best threshold.
y_valid (array-like of shape (n_samples,)) – Ground truth (actual) labels for the validation dataset.
betas (list of float) – A list of beta values to consider when calculating the F-beta score. Beta controls the balance between precision and recall.
y_valid_proba (array-like of shape (n_samples,)) – Predicted probabilities for the positive class in the validation dataset. Used to evaluate thresholds.
kfold (bool, optional) – If
True, returns the best threshold for the given score. If False, updates thethresholdattribute in place. Default isFalse.
- Returns:
The best threshold for the given score if
kfoldisTrue, otherwise returnsNone.- Return type:
float or None
- Raises:
ValueError – Raised if input arrays have mismatched dimensions or invalid beta values.
TypeError – Raised if invalid data types are passed for parameters.
Description:
This method identifies the optimal classification threshold for maximizing the F-beta score.
The F-beta score balances precision and recall, with beta determining the relative weight of recall.
Evaluates thresholds ranging from 0 to 1 (with a step size of 0.01) to find the best threshold for each beta value.
Behavior:
Threshold Evaluation:
For each threshold, computes binary predictions and evaluates the resulting F-beta score.
Penalizes thresholds leading to undesirable outcomes, such as excessive false positives compared to true negatives.
K-Fold Mode:
If
kfold=True, returns the optimal threshold without modifying class attributes.
Non K-Fold Mode:
Updates the
self.thresholdattribute with the optimal threshold for the specified score.
Attributes Used:
self.threshold: Stores the optimal threshold for each scoring metric.self.beta: Stores the beta value corresponding to the maximum F-beta score.
Notes:
The method iterates over thresholds and calculates F-beta scores for each beta value, identifying the best-performing threshold.
Penalizes thresholds where false positives exceed true negatives to ensure practical performance.
Designed to support models evaluated on binary classification tasks.
Example:
optimal_threshold = tune_threshold_Fbeta( score="roc_auc", y_valid=y_valid, betas=[0.5, 1, 2], y_valid_proba=model.predict_proba(X_valid)[:, 1], kfold=False, )
train_val_test_split()
- train_val_test_split(X, y, stratify_y=None, train_size=0.6, validation_size=0.2, test_size=0.2, random_state=3, stratify_cols=None)
Splits data into train, validation, and test sets, supporting stratification by specific columns or the target variable.
- Parameters:
X (
pandas.DataFrameor array-like) – The feature matrix to split.y (
pandas.Seriesor array-like) – The target vector corresponding toX.stratify_y (
pandas.Seriesor None, optional) – Specifies whether to stratify based on the target variable. Default isNone.train_size (float, optional) – Proportion of the data to allocate to the training set. Default is
0.6.validation_size (float, optional) – Proportion of the data to allocate to the validation set. Default is
0.2.test_size (float, optional) – Proportion of the data to allocate to the test set. Default is
0.2.random_state (int, optional) – Random seed for reproducibility. Default is
3.stratify_cols (list,
pandas.DataFrame, or None, optional) – Columns to use for stratification, in addition to or instead ofy. Default isNone.
- Returns:
A tuple containing train, validation, and test sets: (
X_train,X_valid,X_test,y_train,y_valid,y_test).- Return type:
tuple of (
pandas.DataFrame,pandas.Series)- Raises:
ValueError – Raised if the sizes for train, validation, and test do not sum to 1.0 or if invalid stratification keys are provided.
Description:
This function splits data into three sets: train, validation, and test.
Supports stratification based on the target variable (
y) or specific columns (stratify_cols).Ensures the proportions of the split sets are consistent with the specified
train_size,validation_size, andtest_size.
Behavior:
Combines
stratify_colsandy(if both are provided) to create a stratification key.Handles missing values in
stratify_colsby filling with empty strings.Uses a two-step splitting approach:
Splits data into train and combined validation-test sets.
Further splits the combined set into validation and test sets.
Attributes Used:
Handles configurations for stratification and proportional splitting.
Note
The sum of
train_size,validation_size, andtest_sizemust equal1.0.Stratification ensures the distribution of classes or categories is preserved across splits.
The function works seamlessly with both
pandas.DataFrameand array-like data structures.
Example:
X_train, X_valid, X_test, y_train, y_valid, y_test = train_val_test_split( X=features, y=target, stratify_y=target, train_size=0.6, validation_size=0.20, test_size=0.20, random_state=42, stratify_cols=['category_column'] )
get_best_score_params()
- get_best_score_params(X, y)
Retrieves the best hyperparameters for the model based on cross-validation scores for specified metrics.
- Parameters:
X (
pandas.DataFrameor array-like) – The feature matrix for training during hyperparameter tuning.y (
pandas.Seriesor array-like) – The target vector corresponding toX.
- Returns:
None. Updates the class attributes with the best parameters and scores.- Return type:
None- Raises:
ValueError – Raised if
self.gridorself.kfis not properly configured.KeyError – Raised if scoring metrics are missing or invalid.
Description:
This method performs hyperparameter tuning using either grid search, randomized grid search, or Bayesian search.
Identifies the best parameter set for each scoring metric specified in the class’s
scoringattribute.Updates the class attributes with the best estimator and scores.
Supported Search Methods:
Grid Search: Exhaustively searches over all parameter combinations.
Randomized Grid Search: Randomly samples a subset of parameter combinations.
Bayesian Search: Uses Bayesian optimization for hyperparameter tuning.
Behavior:
Randomized Search:
If
self.randomized_gridisTrue, uses`RandomizedSearchCV`to perform hyperparameter tuning.
Bayesian Search:
If
self.bayesianisTrue, usesBayesSearchCVfor Bayesian optimization.Removes any
bayes__prefixed parameters from the grid and uses them as additional arguments forBayesSearchCV.
Grid Search:
Defaults to
GridSearchCVif neitherrandomized_gridnorbayesianis enabled.
After fitting the model:
Updates
self.estimatorandself.test_modelwith the best estimator.Stores the best parameters and score for each scoring metric in
self.best_params_per_score.
Attributes Updated:
self.estimator: Updated with the best model after tuning.self.test_model: Updated with the same best model.self.best_params_per_score: A dictionary storing the best parameters and scores for each scoring metric.
Output:
Prints:
The best parameter set and score for each metric.
A summary of grid scores for all parameter combinations.
Updates class attributes with the tuning results.
Note
Supports custom scoring metrics via
self.custom_scorer.The method assumes
self.kfis a valid cross-validator (e.g.,KFoldorStratifiedKFold) andself.gridis properly defined.Designed to work seamlessly with classification and regression models.
conf_mat_class_kfold()
- conf_mat_class_kfold(X, y, test_model, score=None)
Generates and averages confusion matrices across k-folds, producing a combined classification report.
- Parameters:
X (
pandas.DataFrameor array-like) – The feature matrix for k-fold cross-validation.y (
pandas.Seriesor array-like) – The target vector corresponding toX.test_model (object) – The model to be trained and evaluated on each fold.
score (str, optional) – Optional scoring metric label for reporting purposes. Default is
None.
- Returns:
A dictionary containing the averaged classification report and confusion matrix.
- Return type:
- Raises:
ValueError – Raised if the input data is incompatible with k-fold splitting.
Description:
This method performs k-fold cross-validation to generate confusion matrices for each fold.
Averages the confusion matrices across all folds and produces a combined classification report.
Prints the averaged confusion matrix and classification report.
Behavior:
For each fold in k-fold cross-validation:
Splits the data into training and testing subsets.
Fits the
test_modelon the training subset.Predicts the target values for the testing subset.
Computes the confusion matrix for the fold and appends it to a list.
Aggregates predictions and true labels across all folds.
Averages the confusion matrices and generates an overall classification report.
Output:
Prints:
The averaged confusion matrix across all folds.
The overall classification report across all folds.
Returns:
A dictionary containing:
"Classification Report": The averaged classification report as a dictionary."Confusion Matrix": The averaged confusion matrix as a NumPy array.
Note
Designed for classification tasks evaluated with k-fold cross-validation.
Handles both
pandas.DataFrameand array-like structures forXandy.If
scoreis provided, it is included in the printed report headers.
regression_report_kfold()
- regression_report_kfold(X, y, test_model, score=None)
Generates averaged regression metrics across k-folds.
- Parameters:
X (
pandas.DataFrameor array-like) – The feature matrix for k-fold cross-validation.y (
pandas.Seriesor array-like) – The target vector corresponding toX.test_model (object) – The model to be trained and evaluated on each fold.
score (str, optional) – Optional scoring metric label for reporting purposes. Default is
None.
- Returns:
A dictionary containing averaged regression metrics across all folds.
- Return type:
- Raises:
ValueError – Raised if the input data is incompatible with k-fold splitting.
Description:
This method evaluates regression performance metrics using k-fold cross-validation.
Trains the
test_modelon training splits and evaluates it on validation splits for each fold.Aggregates regression metrics from all folds and calculates their averages.
Behavior:
For each fold in k-fold cross-validation:
Splits the data into training and testing subsets.
Fits the
test_modelon the training subset.Predicts the target values for the testing subset.
Computes regression metrics (e.g., RMSE, MAE, R²) and stores them.
Aggregates metrics across all folds and calculates their mean.
Output:
Prints:
The averaged regression metrics across all folds.
Returns:
A dictionary containing the averaged regression metrics.
Attributes Used:
self.regression_report(): Used to compute regression metrics for each fold.
Note
Designed specifically for regression tasks evaluated with k-fold cross-validation.
Handles both
pandas.DataFrameand array-like structures forXandy.
regression_report()
- regression_report(y_true, y_pred, print_results=True)
Generates a regression report with metrics like Mean Absolute Error, R-squared, and Root Mean Squared Error.
- Parameters:
y_true (array-like) – Ground truth (actual) values for the target variable.
y_pred (array-like) – Predicted values for the target variable.
print_results (bool, optional) – Whether to print the regression metrics to the console. Default is
True.
- Returns:
A dictionary containing various regression metrics.
- Return type:
- Raises:
ValueError – Raised if
y_trueandy_predhave mismatched dimensions.
Description:
Computes common regression metrics to evaluate the performance of a regression model.
Metrics include R², explained variance, mean absolute error (MAE), median absolute error, mean squared error (MSE), and root mean squared error (RMSE).
Metrics Computed:
R²: Coefficient of determination, indicating the proportion of variance in the dependent variable explained by the independent variable(s).Explained Variance: Measures the proportion of variance explained by the model.Mean Absolute Error (MAE): Average of the absolute differences between actual and predicted values.Median Absolute Error: Median of the absolute differences between actual and predicted values.Mean Squared Error (MSE): Average of the squared differences between actual and predicted values.Root Mean Squared Error (RMSE): Square root of the mean squared error.
Behavior:
Computes all metrics and stores them in a dictionary.
Optionally prints the metrics to the console, formatted for easy readability.
Output:
Prints:
A formatted list of regression metrics if
print_results=True.
Returns:
A dictionary containing the computed metrics.
Note
This method is designed for regression tasks and is not applicable to classification models.
The returned dictionary can be used for further analysis or logging.
report_model_metrics()
- report_model_metrics(model, X_valid=None, y_valid=None, threshold=0.5, print_results=True, print_per_fold=False)
Generate a DataFrame of model performance metrics, adapting to regression, binary classification, or multiclass classification problems.
Key Features:
Handles regression, binary classification, and multiclass classification tasks.
Supports K-Fold cross-validation with optional metrics printing for individual folds.
Adapts metrics calculation based on the model’s
model_typeattribute.
- Parameters:
model (object) – The trained model with the necessary attributes and methods for prediction, including
predict_probaorpredict, and attributes likemodel_typeandmulti_label(for multiclass classification).X_valid (pandas.DataFrame or array-like, optional) – Feature set used for validation. If performing K-Fold validation, this represents the entire dataset. Default is
None.y_valid (pandas.Series or array-like, optional) –
Truelabels for the validation dataset. If performing K-Fold validation, this corresponds to the entire dataset. Default isNone.threshold (float, optional) – Classification threshold for binary classification models. Predictions above this threshold are classified as the positive class. Default is
0.5.print_results (bool, optional) – Whether to print the metrics report. Default is
True.print_per_fold (bool, optional) – If performing K-Fold validation, specifies whether to print metrics for each fold. Default is
False.
- Return type:
pandas.DataFrame
- Raises:
ValueError – Raised if the provided
model_typeis invalid or incompatible with the data.AttributeError – Raised if the required attributes or methods are missing from the model.
TypeError – Raised for incorrect parameter types, such as non-numeric thresholds.
- Returns:
A pandas DataFrame containing calculated performance metrics. The structure of the DataFrame depends on the model type:
Regression: Includes Mean Absolute Error (MAE), Mean Squared Error (MSE), Root Mean Squared Error (RMSE), R² Score, and Explained Variance.
Binary Classification: Includes Precision (PPV), Average Precision, Sensitivity, Specificity, AUC-ROC, and Brier Score.
Multiclass Classification: Includes Precision, Recall, and F1-Score for each class, along with weighted averages and accuracy.
Note
For regression models, standard regression metrics are calculated.
For binary classification models, threshold-based metrics are computed using probabilities from
predict_proba.For multiclass classification models, metrics are calculated for each class, along with weighted averages.
K-Fold cross-validation aggregates metrics across folds, with an option to print results for each fold.
Examples:
## Example for binary classification: metrics_df = report_model_metrics(model, X_valid=X_test, y_valid=y_test, threshold=0.5) ## Example for regression: metrics_df = report_model_metrics(model, X_valid=X_test, y_valid=y_test) ## Example for K-Fold validation: metrics_df = report_model_metrics(model, X_valid=X, y_valid=y, print_per_fold=True)
find_optimal_threshold_beta()
- find_optimal_threshold_beta(y, y_proba, target_metric=None, target_score=None, beta_value_range=np.linspace(0.01, 4, 400))
Determine the optimal threshold and beta value for classification models by iteratively tuning the decision boundary to meet a target precision or recall score.
Key Features:
Iteratively finds an optimal threshold using an expanding delta range if an exact match isn’t found.
Supports tuning based on precision or recall.
Uses a progressive delta expansion strategy to allow some flexibility in finding an optimal threshold.
Terminates with a warning if delta exceeds 0.2.
- Parameters:
y (numpy.ndarray or list) – The true labels of the dataset, expected to be binary (0 or 1).
y_proba (numpy.ndarray or list) – The predicted probabilities outputted by a model for the positive class.
target_metric (str, optional) – The performance metric to optimize for. Supports either
"precision"or"recall".target_score (float, optional) – The desired precision or recall score that the function attempts to achieve.
beta_value_range (numpy.ndarray, optional) – The range of beta values to test, controlling the balance between precision and recall. Defaults to an array of 400 values between
0.01and4.- Raises:
ValueError – If
target_metricis not one of"precision"or"recall".Exception – If delta exceeds 0.2, meaning an optimal threshold could not be found within tolerance.
- Return type:
tuple or None
- Returns:
A tuple containing:
threshold (float): The optimal decision threshold for classification.
beta (float): The beta value at which the optimal threshold was found.
Returns
Noneif no suitable threshold is found within the acceptable delta range.Note
Uses the
threshold_tunefunction internally to determine the best threshold for each beta.If no exact match is found initially, the function gradually increases
deltain increments of0.01.If delta exceeds
0.2, the function raises an exception indicating failure to find an optimal threshold.Examples:
## Example usage for optimizing precision threshold, beta = find_optimal_threshold_beta( y=y_test, y_proba=y_probabilities, target_metric="precision", target_score=0.8 ) ## Example usage for optimizing recall threshold, beta = find_optimal_threshold_beta( y=y_test, y_proba=y_probabilities, target_metric="recall", target_score=0.85 ) ## Example handling a case where no exact match is found try: threshold, beta = find_optimal_threshold_beta( y=y_test, y_proba=y_probabilities, target_metric="precision", target_score=0.95 ) except Exception as e: print(f"Could not find an optimal threshold: {e}")
Helper Functions
kfold_split()
- kfold_split(classifier, X, y, stratify=False, scoring=['roc_auc'], n_splits=10, random_state=3)
Splits data using k-fold or stratified k-fold cross-validation.
- Parameters:
classifier (object) – The classifier or model to be evaluated during cross-validation.
X (pandas.DataFrame or array-like) – Features dataset to split into k-folds.
y (pandas.Series or array-like) – Target dataset corresponding to
X.stratify (bool, optional) – Whether to use stratified k-fold cross-validation. If
True, usesStratifiedKFold. Otherwise, usesKFold. Default isFalse.scoring (list of str, optional) – Scoring metric(s) to evaluate during cross-validation. Default is
["roc_auc"].n_splits (int, optional) – Number of splits/folds to create for cross-validation. Default is
10.random_state (int, optional) – Random seed for reproducibility. Default is
3.
- Returns:
A
KFoldorStratifiedKFoldcross-validator object based on thestratifyparameter.- Return type:
sklearn.model_selection.KFoldorsklearn.model_selection.StratifiedKFold- Raises:
ValueError – Raised if invalid parameters (e.g., negative
n_splits) are provided.
Note
Use
stratify=Truefor datasets where maintaining the proportion of classes in each fold is important.Use
stratify=Falsefor general k-fold splitting.
get_cross_validate()
- get_cross_validate(classifier, X, y, kf, scoring=['roc_auc'])
Performs cross-validation using the provided classifier, dataset, and cross-validation strategy.
- Parameters:
classifier (object) – The classifier or model to be evaluated during cross-validation.
X (
pandas.DataFrameor array-like) – Features dataset to use during cross-validation.y (
pandas.Seriesor array-like) – Target dataset corresponding toX.kf (
sklearn.model_selection.KFoldorsklearn.model_selection.StratifiedKFold) – Cross-validator object, such asKFoldorStratifiedKFold, specifying the cross-validation strategy.scoring (list of str, optional) – Scoring metric(s) to evaluate during cross-validation. Default is
["roc_auc"].
- Returns:
A dictionary containing cross-validation results, including train and test scores for each fold.
Returned Dictionary Keys:
test_score: Test scores for each fold.train_score: Training scores for each fold.estimator: The estimator fitted on each fold.fit_time: Time taken to fit the model on each fold.score_time: Time taken to score the model on each fold.
- Return type:
- Raises:
ValueError – Raised if invalid
kforscoringparameters are provided.
Note
Supports multiple scoring metrics, which can be specified as a list (e.g.,
["accuracy", "roc_auc"]).Returns additional information such as train scores and estimators for further analysis.
Ensure the classifier supports the metrics defined in the
scoringparameter.
_confusion_matrix_print()
- _confusion_matrix_print(conf_matrix, labels)
Prints the formatted confusion matrix for binary classification.
- Parameters:
conf_matrix (numpy.ndarray or array-like) – The confusion matrix to print, typically a 2x2 numpy array or similar structure.
labels (list of str) – A list of labels corresponding to the confusion matrix entries in the order
[TN, FP, FN, TP].
Description:
Formats and prints a binary classification confusion matrix with labeled cells for True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN).
Includes additional formatting to enhance readability, such as aligned columns and labeled rows.
Output:
The function prints a structured table representation of the confusion matrix directly to the console.
print_pipeline()
- print_pipeline(pipeline)
Displays an ASCII representation of the pipeline steps for visual clarity.
- Parameters:
pipeline (
sklearn.pipeline.Pipelineor object with astepsattribute) – The pipeline object containing different steps to display. Typically, asklearn.pipeline.Pipelineobject or similar structure.
Description:
This function iterates over the steps in a pipeline and displays each step in a visually formatted ASCII art representation.
For each pipeline step:
Displays the step name and its class name in a boxed format.
Connects steps with vertical connectors (│) and arrows (▼) for clarity.
Dynamically adjusts box width based on the longest step name or class name to maintain alignment.
Output:
The function prints the pipeline structure directly to the console, providing an easy-to-read ASCII visualization.
Note
If the pipeline has no steps or lacks a
stepsattribute, the function prints a message:"No steps found in the pipeline!".Designed for readability, especially in terminal environments.
Pipeline Management
The pipeline in the model tuner class is designed to automatically organize steps into three categories: preprocessing, feature selection, and imbalanced sampling. The steps are ordered in the following sequence:
Preprocessing:
Imputation
Scaling
Other preprocessing steps
Imbalanced Sampling
Feature Selection
Classifier
The pipeline_assembly method automatically sorts the steps into this order.
Specifying Pipeline Steps
Pipeline steps can be specified in multiple ways. For example, if naming a pipeline step then specify like so:
pipeline_steps = ['imputer', SimpleImputer()]
Naming each step is optional and the steps can also be specified like so:
pipeline_steps = [SimpleImputer(), StandardScalar(), rfe()]
If no name is assigned, the step will be renamed automatically to follow the convention
step_0,step_1, etc.Column transformers can also be included in the pipeline and are automatically categorized under the preprocessing section.
Helper Methods for Pipeline Extraction
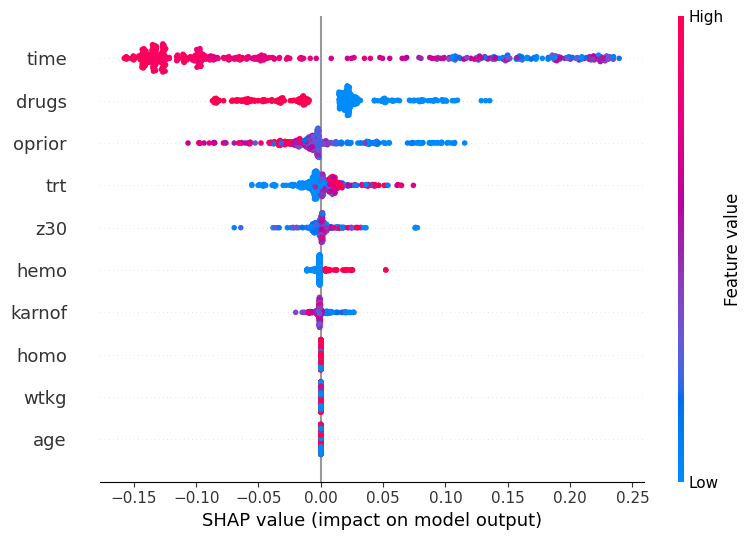
To support advanced use cases, the model tuner provides helper methods to extract parts of the pipeline for later use. For example, when generating SHAP plots, users might only need the preprocessing section of the pipeline.
Here are some of the available methods:
- get_preprocessing_and_feature_selection_pipeline()
Extracts both the preprocessing and feature selection parts of the pipeline.
Definition:
def get_preprocessing_and_feature_selection_pipeline(self): steps = [ (name, transformer) for name, transformer in self.estimator.steps if name.startswith("preprocess_") or name.startswith("feature_selection_") ] return self.PipelineClass(steps)
- get_feature_selection_pipeline()
Extracts only the feature selection part of the pipeline.
Definition:
def get_feature_selection_pipeline(self): steps = [ (name, transformer) for name, transformer in self.estimator.steps if name.startswith("feature_selection_") ] return self.PipelineClass(steps)
- get_preprocessing_pipeline()
Extracts only the preprocessing part of the pipeline.
Definition:
def get_preprocessing_pipeline(self): preprocessing_steps = [ (name, transformer) for name, transformer in self.estimator.steps if name.startswith("preprocess_") ] return self.PipelineClass(preprocessing_steps)
Extracting Feature names
When performing feature selection with tools such as Recursive Feature Elimination (RFE) or when using ColumnTransformers the feature names that are fed to the model can be obscured and different from the original. To get the transformed feature names or to extract the feature names that were selected by the feature selection process we have provided the get_feature_names() method.
- get_feature_names()
Extracts the feature names after they have been processed by the pipeline. This does not work if a ColumnTransformerm, OneHotEncoder or some form of feature selection is not present in the pipeline.
Definition:
def get_feature_names(self): if self.pipeline_steps is None or not self.pipeline_steps: raise ValueError("You must provide pipeline steps to use get_feature_names") if hasattr(self.estimator, "steps"): estimator_steps = self.estimator[:-1] else: estimator_steps = self.estimator.estimator[:-1] return estimator_steps.get_feature_names_out().tolist()
Example Usage:
### Assuming you already have fitted a model with some form of feature selection ### or feature transformation in the pipeline e.g. one hot encoder: feat_names = model.get_feature_names()
Summary
By organizing pipeline steps automatically and providing helper methods for extraction, the model tuner class offers flexibility and ease of use for building and managing complex pipelines. Users can focus on specifying the steps, and the tuner handles naming, sorting, and category assignments seamlessly.
Binary Classification
Binary classification is a type of supervised learning where a model is trained
to distinguish between two distinct classes or categories. In essence, the model
learns to classify input data into one of two possible outcomes, typically
labeled as 0 and 1, or negative and positive. This is commonly used in
scenarios such as spam detection, disease diagnosis, or fraud detection.
The model_tuner library handles binary classification seamlessly through the Model
class. Users can specify a binary classifier as the estimator, and the library
takes care of essential tasks like data preprocessing, model calibration, and
cross-validation. The library also provides robust support for evaluating the
model’s performance using a variety of metrics, such as accuracy, precision,
recall, and ROC-AUC, ensuring that the model’s ability to distinguish between the
two classes is thoroughly assessed. Additionally, the library supports advanced
techniques like imbalanced data handling and model calibration to fine-tune
decision thresholds, making it easier to deploy effective binary classifiers in
real-world applications.
AIDS Clinical Trials Group Study
The UCI Machine Learning Repository is a well-known resource for accessing a wide range of datasets used for machine learning research and practice. One such dataset is the AIDS Clinical Trials Group Study dataset, which can be used to build and evaluate predictive models.
You can easily fetch this dataset using the ucimlrepo package. If you haven’t installed it yet, you can do so by running the following command:
pip install ucimlrepo
Once installed, you can quickly load the AIDS Clinical Trials Group Study dataset with a simple command:
from ucimlrepo import fetch_ucirepo
Step 1: Import Necessary libraries
import pandas as pd
import numpy as np
from ucimlrepo import fetch_ucirepo
from xgboost import XGBClassifier
from model_tuner import Model
Step 2: Load the dataset, define X, y
## Fetch dataset
aids_clinical_trials_group_study_175 = fetch_ucirepo(id=890)
## Data (as pandas dataframes)
X = aids_clinical_trials_group_study_175.data.features
y = aids_clinical_trials_group_study_175.data.targets
y = y.squeeze() ## convert a DataFrame to Series when single column
Step 3: Check for zero-variance columns and drop accordingly
## Check for zero-variance columns and drop them
zero_variance_columns = X.columns[X.var() == 0]
if not zero_variance_columns.empty:
X = X.drop(columns=zero_variance_columns)
Step 4: Create an instance of the XGBClassifier
## Creating an instance of the XGBClassifier
xgb_name = "xgb"
xgb = XGBClassifier(
objective="binary:logistic",
random_state=222,
)
Step 5: Define Hyperparameters for XGBoost
In binary classification, we configure the XGBClassifier for tasks where the
model predicts between two classes (e.g., positive/negative or 0/1). Here, we
define a grid of hyperparameters to fine-tune the XGBoost model.
The following code defines the hyperparameter grid and configuration:
xgbearly = True
tuned_parameters_xgb = {
f"{xgb_name}__max_depth": [3, 10, 20, 200, 500],
f"{xgb_name}__learning_rate": [1e-4],
f"{xgb_name}__n_estimators": [1000],
f"{xgb_name}__early_stopping_rounds": [100],
f"{xgb_name}__verbose": [0],
f"{xgb_name}__eval_metric": ["logloss"],
}
## Define model configuration
xgb_definition = {
"clc": xgb,
"estimator_name": xgb_name,
"tuned_parameters": tuned_parameters_xgb,
"randomized_grid": False,
"n_iter": 5, ## Number of iterations if randomized_grid=True
"early": xgbearly,
}
Key Configurations
Hyperparameter Grid:
max_depth: Limits the depth of each decision tree to prevent overfitting.learning_rate: Controls the impact of each boosting iteration; smaller values require more boosting rounds.n_estimators: Specifies the total number of boosting rounds.verbose: Controls output during training; set to0for silent mode or1to display progress.eval_metric: Measures model performance (e.g.,loglossfor binary classification), evaluating the negative log-likelihood.early_stopping_rounds: Halts training early if validation performance does not improve after the specified number of rounds.
General Settings:
Use
randomized_grid=Falseto perform exhaustive grid search.Set the number of iterations for randomized search with
n_iterif needed.
The grid search will explore the parameter combinations to find the optimal configuration for binary classification tasks.
Note
The verbose parameter in XGBoost allows you to control the level of output during training:
Set to
0orFalse: Suppresses all training output (silent mode).Set to
1orTrue: Displays progress and evaluation metrics during training.
This can be particularly useful for monitoring model performance when early stopping is enabled.
Important
When defining hyperparameters for boosting algorithms, frameworks like
XGBoost allow straightforward configuration, such as specifying n_estimators
to control the number of boosting rounds. However, CatBoost introduces certain
pitfalls when this parameter is defined.
Refer to the important caveat regarding this scenario for further details.
Step 6: Initialize and configure the Model
XGBClassifier inherently handles missing values (NaN) without requiring explicit
imputation strategies. During training, XGBoost treats missing values as a
separate category and learns how to route them within its decision trees.
Therefore, passing a SimpleImputer or using an imputation strategy is unnecessary
when using XGBClassifier.
model_type = "xgb"
clc = xgb_definition["clc"]
estimator_name = xgb_definition["estimator_name"]
tuned_parameters = xgb_definition["tuned_parameters"]
n_iter = xgb_definition["n_iter"]
rand_grid = xgb_definition["randomized_grid"]
early_stop = xgb_definition["early"]
kfold = False
calibrate = True
## Initialize model_tuner
model_xgb = Model(
name=f"AIDS_Clinical_{model_type}",
estimator_name=estimator_name,
calibrate=calibrate,
estimator=clc,
model_type="classification",
kfold=kfold,
stratify_y=True,
stratify_cols=["gender", "race"],
grid=tuned_parameters,
randomized_grid=rand_grid,
boost_early=early_stop,
scoring=["roc_auc"],
random_state=222,
n_jobs=2,
)
Step 7: Perform grid search parameter tuning and retrieve split data
## Perform grid search parameter tuning
model_xgb.grid_search_param_tuning(X, y, f1_beta_tune=True)
## Get the training, validation, and test data
X_train, y_train = model_xgb.get_train_data(X, y)
X_valid, y_valid = model_xgb.get_valid_data(X, y)
X_test, y_test = model_xgb.get_test_data(X, y)
With the model configured, the next step is to perform grid search parameter tuning
to find the optimal hyperparameters for the XGBClassifier. The
grid_search_param_tuning method will iterate over all combinations of
hyperparameters specified in tuned_parameters, evaluate each one using the
specified scoring metric, and select the best performing set.
In this example, we pass an additional argument, f1_beta_tune=True, which
adjusts the F1 score to weigh precision and recall differently during
hyperparameter optimization.
Note
For a more in depth discussion on threshold tuning, refer to this section.
Why use f1_beta_tune=True?
Standard F1-Score: Balances precision and recall equally (
beta=1).Custom Beta Values:
With
f1_beta_tune=True, the model tunes the decision threshold to optimize a custom F1 score using the beta value specified internally.This is useful in scenarios where one metric (precision or recall) is more critical than the other.
This method will:
Split the Data: The data will be split into training and validation sets. Since
stratify_y=True, the class distribution will be maintained across splits.After tuning, retrieve the training, validation, and test splits using:
get_train_datafor training data.get_valid_datafor validation data.get_test_datafor test data.
Iterate Over Hyperparameters: All combinations of hyperparameters defined in
tuned_parameterswill be tried sincerandomized_grid=False.Early Stopping: With
boost_early=Trueandearly_stopping_roundsset in the hyperparameters, the model will stop training early if the validation score does not improve.Optimize for Scoring Metric: The model uses
roc_auc(ROC AUC) as the scoring metric suitable for binary classification.Select Best Model: The hyperparameter set that yields the best validation score will be selected.
Pipeline Steps:
┌─────────────────┐
│ Step 1: xgb │
│ XGBClassifier │
└─────────────────┘
100%|██████████| 5/5 [00:47<00:00, 9.43s/it]
Fitting model with best params and tuning for best threshold ...
100%|██████████| 2/2 [00:00<00:00, 2.87it/s]Best score/param set found on validation set:
{'params': {'xgb__early_stopping_rounds': 100,
'xgb__eval_metric': 'logloss',
'xgb__learning_rate': 0.0001,
'xgb__max_depth': 3,
'xgb__n_estimators': 999},
'score': 0.9260891500474834}
Best roc_auc: 0.926
Step 8: Fit the model
In this step, we train the XGBClassifier using the training data and monitor
performance on the validation data during training.
model_xgb.fit(
X_train,
y_train,
validation_data=[X_valid, y_valid],
)
Note
The inclusion of validation_data allows XGBoost to:
Monitor Validation Performance: XGBoost evaluates the model’s performance on the validation set after each boosting round using the specified evaluation metric (e.g.,
logloss).Enable Early Stopping: If
early_stopping_roundsis defined, training will stop automatically if the validation performance does not improve after a set number of rounds, preventing overfitting and saving computation time.
Step 9: Return metrics (optional)
Hint
Use the return metrics function to evaluate the model by printing the output.
# ------------------------- VALID AND TEST METRICS -----------------------------
print("Validation Metrics")
model_xgb.return_metrics(
X=X_valid,
y=y_valid,
optimal_threshold=True,
print_threshold=True,
model_metrics=True,
)
print()
print("Test Metrics")
model_xgb.return_metrics(
X=X_test,
y=y_test,
optimal_threshold=True,
print_threshold=True,
model_metrics=True,
)
print()
Validation Metrics
Confusion matrix on set provided:
--------------------------------------------------------------------------------
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 93 (tp) 11 (fn)
Neg 76 (fp) 248 (tn)
--------------------------------------------------------------------------------
********************************************************************************
Report Model Metrics: xgb
Metric Value
0 Precision/PPV 0.550296
1 Average Precision 0.802568
2 Sensitivity 0.894231
3 Specificity 0.765432
4 AUC ROC 0.926089
5 Brier Score 0.166657
********************************************************************************
--------------------------------------------------------------------------------
precision recall f1-score support
0 0.96 0.77 0.85 324
1 0.55 0.89 0.68 104
accuracy 0.80 428
macro avg 0.75 0.83 0.77 428
weighted avg 0.86 0.80 0.81 428
--------------------------------------------------------------------------------
Optimal threshold used: 0.26
Test Metrics
Confusion matrix on set provided:
--------------------------------------------------------------------------------
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 99 (tp) 6 (fn)
Neg 82 (fp) 241 (tn)
--------------------------------------------------------------------------------
********************************************************************************
Report Model Metrics: xgb
Metric Value
0 Precision/PPV 0.546961
1 Average Precision 0.816902
2 Sensitivity 0.942857
3 Specificity 0.746130
4 AUC ROC 0.934306
5 Brier Score 0.167377
********************************************************************************
--------------------------------------------------------------------------------
precision recall f1-score support
0 0.98 0.75 0.85 323
1 0.55 0.94 0.69 105
accuracy 0.79 428
macro avg 0.76 0.84 0.77 428
weighted avg 0.87 0.79 0.81 428
--------------------------------------------------------------------------------
Optimal threshold used: 0.26
Note
A detailed classification report is also available at this stage for review. To print and examine it, refer to this Model Metrics section for guidance on accessing and interpreting the report.
Step 10: Calibrate the model (if needed)
See this section for more information on model calibration.
import matplotlib.pyplot as plt
from sklearn.calibration import calibration_curve
## Get the predicted probabilities for the validation data from uncalibrated model
y_prob_uncalibrated = model_xgb.predict_proba(X_test)[:, 1]
## Compute the calibration curve for the uncalibrated model
prob_true_uncalibrated, prob_pred_uncalibrated = calibration_curve(
y_test,
y_prob_uncalibrated,
n_bins=10,
)
## Calibrate the model
if model_xgb.calibrate:
model_xgb.calibrateModel(X, y, score="roc_auc")
## Predict on the validation set
y_test_pred = model_xgb.predict_proba(X_test)[:, 1]
Change back to CPU
Confusion matrix on validation set for roc_auc
--------------------------------------------------------------------------------
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 74 (tp) 30 (fn)
Neg 20 (fp) 304 (tn)
--------------------------------------------------------------------------------
precision recall f1-score support
0 0.91 0.94 0.92 324
1 0.79 0.71 0.75 104
accuracy 0.88 428
macro avg 0.85 0.82 0.84 428
weighted avg 0.88 0.88 0.88 428
--------------------------------------------------------------------------------
roc_auc after calibration: 0.9260891500474834
## Get the predicted probabilities for the validation data from calibrated model
y_prob_calibrated = model_xgb.predict_proba(X_test)[:, 1]
## Compute the calibration curve for the calibrated model
prob_true_calibrated, prob_pred_calibrated = calibration_curve(
y_test,
y_prob_calibrated,
n_bins=10,
)
## Plot the calibration curves
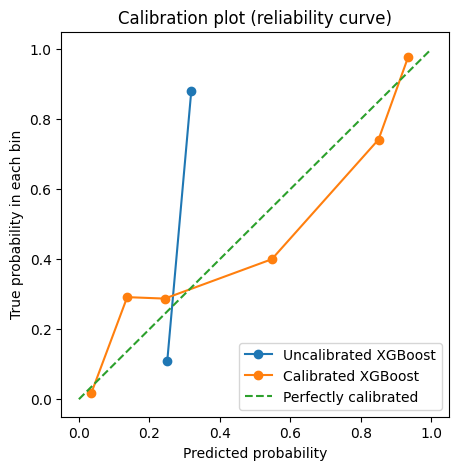
plt.figure(figsize=(5, 5))
plt.plot(
prob_pred_uncalibrated,
prob_true_uncalibrated,
marker="o",
label="Uncalibrated XGBoost",
)
plt.plot(
prob_pred_calibrated,
prob_true_calibrated,
marker="o",
label="Calibrated XGBoost",
)
plt.plot(
[0, 1],
[0, 1],
linestyle="--",
label="Perfectly calibrated",
)
plt.xlabel("Predicted probability")
plt.ylabel("True probability in each bin")
plt.title("Calibration plot (reliability curve)")
plt.legend()
plt.show()
F1 Beta Threshold Tuning
In binary classification, selecting an optimal classification threshold is crucial for achieving the best balance between precision and recall. The default threshold of 0.5 may not always yield optimal performance, especially when dealing with imbalanced datasets. F1 Beta threshold tuning helps adjust this threshold to maximize the F-beta score, which balances precision and recall according to the importance assigned to each through the beta parameter.
Note
To better understand the impact of threshold tuning on model results, see the Threshold Tuning Considerations section.
Understanding F1 Beta Score
The F-beta score is a generalization of the F1-score that allows you to weigh recall more heavily than precision (or vice versa) based on the beta parameter:
F1-Score (beta = 1): Equal importance to precision and recall.
F-beta > 1: More emphasis on recall. Useful when false negatives are more critical (e.g., disease detection).
F-beta < 1: More emphasis on precision. Suitable when false positives are costlier (e.g., spam detection).
Example usage: default (beta = 1)
Setting up the Model object ready for tuning.
from xgboost import XGBClassifier
xgb_name = "xgb"
xgb = XGBClassifier(
objective="binary:logistic"
random_state=222,
)
tuned_parameters_xgb = {
f"{xgb_name}__max_depth": [3, 10, 20, 200, 500],
f"{xgb_name}__learning_rate": [1e-4],
f"{xgb_name}__n_estimators": [1000],
f"{xgb_name}__early_stopping_rounds": [100],
f"{xgb_name}__verbose": [0],
f"{xgb_name}__eval_metric": ["logloss"],
}
xgb_model = Model(
name=f"Threshold Example Model",
estimator_name=xgb_name,
calibrate=False,
model_type="classification",
estimator=xgb,
kfold=False,
stratify_y=True,
stratify_cols=False,
grid=tuned_parameters_xgb,
randomized_grid=False,
boost_early=False,
scoring=["roc_auc"],
random_state=222,
n_jobs=2,
)
In the grid_search_param_tuning use the f1_beta_tune variable when using
grid_search_param_tuning(). Set this to True to enable tuning.
xgb_model.grid_search_param_tuning(X, y, f1_beta_tune=True)
This will find the best hyperparameters and then find the best threshold for these balancing both precision and recall. The threshold is stored in the Model object. To access this:
xgb_model.threshold
This will give the best threshold found for each score specified in the Model object.
When using methods to return metrics or report metrics and an optimal threshold was used make sure to remember to specify this in them for example:
xgb_model.return_metrics(
X_valid,
y_valid,
optimal_threshold=True,
print_threshold=True,
model_metrics=True,
)
Example usage: custom betas (higher recall)
If we want to have a higher recall score and care less about precision then we increase
the beta value. This looks very similar to the previous example except that when we
use f1_beta_tune, we also set a beta value like so:
xgb_model.grid_search_param_tuning(X, y, f1_beta_tune=True, betas=[2])
Setting the beta value to 2 will priortise increasing the recall over the precision.
Example usage: custom betas (higher precision)
If we want to have a higher precision score and care less about recall then we decrease the beta value. This looks very similar to the previous example except that we set the beta value to less than 1.
xgb_model.grid_search_param_tuning(X, y, f1_beta_tune=True, betas=[0.5])
Setting the beta value to 0.5 will priortise increasing the precision over the recall.
Optimizing Model Threshold for Precision/Recall Trade-off
This function helps fine-tune a saved model’s decision threshold to maximize precision or recall using a beta-weighted approach.
Note
See this section for a more detailed explanation with contextual examples.
threshold, beta = find_optimal_threshold_beta(
y_valid,
model.predict_proba(X_valid)[:, 1],
target_metric="precision",
target_score=0.5,
beta_value_range=np.linspace(0.01, 4, 40),
)
Imbalanced Learning
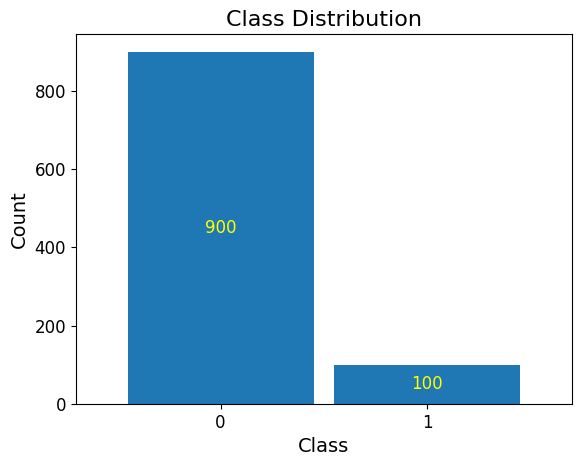
In machine learning, imbalanced datasets are a frequent challenge, especially in real-world scenarios. These datasets have an unequal distribution of target classes, with one class (e.g., fraudulent transactions, rare diseases, or other low-frequency events) being underrepresented compared to the majority class. Models trained on imbalanced data often struggle to generalize, as they tend to favor the majority class, leading to poor performance on the minority class.
To mitigate these issues, it is crucial to:
Understand the nature of the imbalance in the dataset.
Apply appropriate resampling techniques (oversampling, undersampling, or hybrid methods).
Use metrics beyond accuracy, such as precision, recall, and F1-score, to evaluate model performance fairly.
Generating an imbalanced dataset
Demonstrated below are the steps to generate an imbalanced dataset using
make_classification from the sklearn.datasets module. The following
parameters are specified:
n_samples=1000: The dataset contains 1,000 samples.n_features=20: Each sample has 20 features.n_informative=2: Two features are informative for predicting the target.n_redundant=2: Two features are linear combinations of the informative features.weights=[0.9, 0.1]: The target class distribution is 90% for the majority class and 10% for the minority class, creating an imbalance.flip_y=0: No label noise is added to the target variable.random_state=42: Ensures reproducibility by using a fixed random seed.
import pandas as pd
import numpy as np
from sklearn.datasets import make_classification
X, y = make_classification(
n_samples=1000,
n_features=20,
n_informative=2,
n_redundant=2,
n_clusters_per_class=1,
weights=[0.9, 0.1],
flip_y=0,
random_state=42,
)
## Convert to a pandas DataFrame for better visualization
data = pd.DataFrame(X, columns=[f'feature_{i}' for i in range(1, 21)])
data['target'] = y
X = data[[col for col in data.columns if "target" not in col]]
y = pd.Series(data["target"])
Below, you will see that the dataset we have generated is severely imbalanced with 900 observations allocated to the majority class (0) and 100 observations to the minority class (1).
import matplotlib.pyplot as plt
## Create a bar plot
value_counts = pd.Series(y).value_counts()
ax = value_counts.plot(
kind="bar",
rot=0,
width=0.9,
)
## Add labels inside the bars
for index, count in enumerate(value_counts):
plt.text(
index,
count / 2,
str(count),
ha="center",
va="center",
color="yellow",
)
## Customize labels and title
plt.xlabel("Class")
plt.ylabel("Count")
plt.title("Class Distribution")
plt.show() ## Show the plot
Define hyperparameters for XGBoost
Below, we will use an XGBoost classifier with the following hyperparameters:
from xgboost import XGBClassifier
xgb_name = "xgb"
xgb = XGBClassifier(
random_state=222,
)
xgbearly = True
tuned_parameters_xgb = {
f"{xgb_name}__max_depth": [3, 10, 20, 200, 500],
f"{xgb_name}__learning_rate": [1e-4],
f"{xgb_name}__n_estimators": [1000],
f"{xgb_name}__early_stopping_rounds": [100],
f"{xgb_name}__verbose": [0],
f"{xgb_name}__eval_metric": ["logloss"],
}
xgb_definition = {
"clc": xgb,
"estimator_name": xgb_name,
"tuned_parameters": tuned_parameters_xgb,
"randomized_grid": False,
"n_iter": 5,
"early": xgbearly,
}
Define the model object
model_type = "xgb"
clc = xgb_definition["clc"]
estimator_name = xgb_definition["estimator_name"]
tuned_parameters = xgb_definition["tuned_parameters"]
n_iter = xgb_definition["n_iter"]
rand_grid = xgb_definition["randomized_grid"]
early_stop = xgb_definition["early"]
kfold = False
calibrate = True
Addressing Class Imbalance in Machine Learning
Class imbalance occurs when one class significantly outweighs another in the dataset, leading to biased models that perform well on the majority class but poorly on the minority class. Techniques like SMOTE and others aim to address this issue by improving the representation of the minority class, ensuring balanced learning and better generalization.
Techniques to Address Class Imbalance
Resampling Techniques
SMOTE (Synthetic Minority Oversampling Technique): SMOTE generates synthetic samples for the minority class by interpolating between existing minority class data points and their nearest neighbors. This helps create a more balanced class distribution without merely duplicating data, thus avoiding overfitting.
Oversampling: Randomly duplicates examples from the minority class to balance the dataset. While simple, it risks overfitting to the duplicated examples.
Undersampling: Reduces the majority class by randomly removing samples. While effective, it can lead to loss of important information.
Purpose of Using These Techniques
The goal of using these techniques is to improve model performance on imbalanced datasets, specifically by:
Ensuring the model captures meaningful patterns in the minority class.
Reducing bias toward the majority class, which often dominates predictions in imbalanced datasets.
Improving metrics like recall, F1-score, and AUC-ROC for the minority class, which are critical in applications like fraud detection, healthcare, and rare event prediction.
Note
While we provide comprehensive examples for SMOTE, ADASYN, and
RandomUnderSampler in the accompanying notebook,
this documentation section demonstrates the implementation of SMOTE. The other
examples follow a similar workflow and can be executed by simply passing the
respective imbalance_sampler input to ADASYN() or RandomUnderSampler(), as
needed. For detailed examples of all methods, please refer to the linked notebook.
Synthetic Minority Oversampling Technique (SMOTE)
SMOTE (Synthetic Minority Oversampling Technique) is a method used to address class imbalance in datasets. It generates synthetic samples for the minority class by interpolating between existing minority samples and their nearest neighbors, effectively increasing the size of the minority class without duplicating data. This helps models better learn patterns from the minority class, improving classification performance on imbalanced datasets.
Step 1: Initalize and configure the model
Important
In the code block below, we initialize and configure the model by instantiating the
Model class and assigning it to a variable named xgb_smote. Note that
the imbalance_sampler=SMOTE(random_state=42) parameter is included to activate
the imbalanced sampler. Setting a random state of 42 ensures reproducibility of results.
from model_tuner import Model
from imblearn.over_sampling import SMOTE
xgb_smote = Model(
name=f"Make_Classification_{model_type}",
estimator_name=estimator_name,
calibrate=calibrate,
model_type="classification",
estimator=clc,
kfold=kfold,
stratify_y=True,
stratify_cols=False,
grid=tuned_parameters,
randomized_grid=rand_grid,
boost_early=early_stop,
scoring=["roc_auc"],
random_state=222,
n_jobs=2,
imbalance_sampler=SMOTE(random_state=42),
)
Step 2: Perform grid search parameter tuning and retrieve split data
xgb_smote.grid_search_param_tuning(X, y, f1_beta_tune=True)
## Get the training, validation, and test data
X_train, y_train = xgb_smote.get_train_data(X, y)
X_valid, y_valid = xgb_smote.get_valid_data(X, y)
X_test, y_test = xgb_smote.get_test_data(X, y)
Pipeline Steps:
┌─────────────────────┐
│ Step 1: resampler │
│ SMOTE │
└─────────────────────┘
│
▼
┌─────────────────────┐
│ Step 2: xgb │
│ XGBClassifier │
└─────────────────────┘
Distribution of y values after resampling: target
0 540
1 540
Name: count, dtype: int64
100%|██████████| 5/5 [00:16<00:00, 3.25s/it]
Fitting model with best params and tuning for best threshold ...
100%|██████████| 2/2 [00:00<00:00, 4.17it/s]Best score/param set found on validation set:
{'params': {'xgb__early_stopping_rounds': 100,
'xgb__eval_metric': 'logloss',
'xgb__learning_rate': 0.0001,
'xgb__max_depth': 3,
'xgb__n_estimators': 999},
'score': 0.9969444444444445}
Best roc_auc: 0.997
SMOTE: Distribution of y values after resampling
Notice that the target has been redistributed after SMOTE to 540 observations for the minority class and 540 observations for the majority class.
Step 3: Fit the model
xgb_smote.fit(
X_train,
y_train,
validation_data=[X_valid, y_valid],
)
Step 4: Return metrics (optional)
# ------------------------- VALID AND TEST METRICS -----------------------------
print("Validation Metrics")
xgb_smote.return_metrics(
X_valid,
y_valid,
optimal_threshold=True,
print_threshold=True,
model_metrics=True,
)
print()
print("Test Metrics")
xgb_smote.return_metrics(
X_test,
y_test,
optimal_threshold=True,
print_threshold=True,
model_metrics=True,
)
print()
Validation Metrics
Confusion matrix on set provided:
--------------------------------------------------------------------------------
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 20 (tp) 0 (fn)
Neg 1 (fp) 179 (tn)
--------------------------------------------------------------------------------
********************************************************************************
Report Model Metrics: xgb
Metric Value
0 Precision/PPV 0.952381
1 Average Precision 0.947751
2 Sensitivity 1.000000
3 Specificity 0.994444
4 AUC ROC 0.996944
5 Brier Score 0.208997
********************************************************************************
--------------------------------------------------------------------------------
precision recall f1-score support
0 1.00 0.99 1.00 180
1 0.95 1.00 0.98 20
accuracy 0.99 200
macro avg 0.98 1.00 0.99 200
weighted avg 1.00 0.99 1.00 200
--------------------------------------------------------------------------------
Optimal threshold used: 0.52
Test Metrics
Confusion matrix on set provided:
--------------------------------------------------------------------------------
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 18 (tp) 2 (fn)
Neg 3 (fp) 177 (tn)
--------------------------------------------------------------------------------
********************************************************************************
Report Model Metrics: xgb
Metric Value
0 Precision/PPV 0.857143
1 Average Precision 0.897215
2 Sensitivity 0.900000
3 Specificity 0.983333
4 AUC ROC 0.966944
5 Brier Score 0.209358
********************************************************************************
--------------------------------------------------------------------------------
precision recall f1-score support
0 0.99 0.98 0.99 180
1 0.86 0.90 0.88 20
accuracy 0.97 200
macro avg 0.92 0.94 0.93 200
weighted avg 0.98 0.97 0.98 200
--------------------------------------------------------------------------------
Optimal threshold used: 0.52
Recursive Feature Elimination (RFE)
Now that we’ve trained the models, we can also refine them by identifying which features contribute most to their performance. One effective method for this is Recursive Feature Elimination (RFE). This technique allows us to systematically remove the least important features, retraining the model at each step to evaluate how performance is affected. By focusing only on the most impactful variables, RFE helps streamline the dataset, reduce noise, and improve both the accuracy and interpretability of the final model.
It works by recursively training a model, ranking the importance of features based on the model’s outputas (such as coefficients in linear models or importance scores in tree-based models), and then removing the least important features one by one. This process continues until a specified number of features remains or the desired performance criteria are met.
The primary advantage of RFE is its ability to streamline datasets, improving model performance and interpretability by focusing on features that contribute the most to the predictive power. However, it can be computationally expensive since it involves repeated model training, and its effectiveness depends on the underlying model’s ability to evaluate feature importance. RFE is commonly used with cross-validation to ensure that the selected features generalize well across datasets, making it a robust choice for model optimization and dimensionality reduction.
As an illustrative example, we will retrain the above model using RFE.
from sklearn.feature_selection import RFE
from sklearn.linear_model import ElasticNet
We will begin by appending the feature selection technique to our tuned parameters dictionary.
xgb_definition["tuned_parameters"][f"feature_selection_rfe__n_features_to_select"] = [
None,
5,
10,
]
Elastic Net for feature selection with RFE
Note
You may wish to explore this section for the rationale in applying this technique.
We will use elastic net because it strikes a balance between two widely used regularization techniques: Lasso (\(L1\)) and Ridge (\(L2\)). Elastic net is particularly effective in scenarios where we expect the dataset to have a mix of strongly and weakly correlated features. Lasso alone tends to select only one feature from a group of highly correlated ones, ignoring the others, while Ridge includes all features but may not perform well when some are entirely irrelevant. Elastic net addresses this limitation by combining both penalties, allowing it to handle multicollinearity more effectively while still performing feature selection.
Additionally, elastic net provides flexibility by controlling the ratio between \(L1\) and \(L2\) penalties, enabling fine-tuning to suit the specific needs of our dataset. This makes it a robust choice for datasets with many features, some of which may be irrelevant or redundant, as it can reduce overfitting while retaining a manageable subset of predictors.
rfe_estimator = ElasticNet(alpha=10.0, l1_ratio=0.9)
rfe = RFE(rfe_estimator)
from model_tuner import Model
model_xgb = Model(
name=f"AIDS_Clinical_{model_type}",
estimator_name=estimator_name,
calibrate=calibrate,
estimator=clc,
model_type="classification",
kfold=kfold,
pipeline_steps=[
("rfe", rfe),
],
stratify_y=True,
stratify_cols=False,
grid=tuned_parameters,
randomized_grid=rand_grid,
feature_selection=True,
boost_early=early_stop,
scoring=["roc_auc"],
random_state=222,
n_jobs=2,
)
model_xgb.grid_search_param_tuning(X, y, f1_beta_tune=True)
X_train, y_train = model_xgb.get_train_data(X, y)
X_valid, y_valid = model_xgb.get_valid_data(X, y)
X_test, y_test = model_xgb.get_test_data(X, y)
model_xgb.fit(
X_train,
y_train,
validation_data=[X_valid, y_valid],
)
# ------------------------- VALID AND TEST METRICS -----------------------------
print("Validation Metrics")
model_xgb.return_metrics(
X_valid,
y_valid,
optimal_threshold=True,
print_threshold=True,
model_metrics=True,
)
print()
print("Test Metrics")
model_xgb.return_metrics(
X_test,
y_test,
optimal_threshold=True,
print_threshold=True,
model_metrics=True,
)
print()
Pipeline Steps:
┌─────────────────────────────────┐
│ Step 1: feature_selection_rfe │
│ RFE │
└─────────────────────────────────┘
│
▼
┌─────────────────────────────────┐
│ Step 2: xgb │
│ XGBClassifier │
└─────────────────────────────────┘
100%|██████████| 15/15 [00:40<00:00, 2.70s/it]
Fitting model with best params and tuning for best threshold ...
100%|██████████| 2/2 [00:00<00:00, 3.53it/s]
Best score/param set found on validation set:
{'params': {'feature_selection_rfe__n_features_to_select': 10,
'xgb__early_stopping_rounds': 100,
'xgb__eval_metric': 'logloss',
'xgb__learning_rate': 0.0001,
'xgb__max_depth': 10,
'xgb__n_estimators': 999},
'score': 0.9324994064577399}
Best roc_auc: 0.932
Validation Metrics
Confusion matrix on set provided:
--------------------------------------------------------------------------------
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 94 (tp) 10 (fn)
Neg 70 (fp) 254 (tn)
--------------------------------------------------------------------------------
********************************************************************************
Report Model Metrics: xgb
Metric Value
0 Precision/PPV 0.573171
1 Average Precision 0.824825
2 Sensitivity 0.903846
3 Specificity 0.783951
4 AUC ROC 0.932499
5 Brier Score 0.165950
********************************************************************************
--------------------------------------------------------------------------------
precision recall f1-score support
0 0.96 0.78 0.86 324
1 0.57 0.90 0.70 104
accuracy 0.81 428
macro avg 0.77 0.84 0.78 428
weighted avg 0.87 0.81 0.82 428
--------------------------------------------------------------------------------
Feature names selected:
['time', 'preanti', 'str2', 'strat', 'symptom', 'treat', 'offtrt', 'cd40', 'cd420', 'cd80']
Optimal threshold used: 0.25
Test Metrics
Confusion matrix on set provided:
--------------------------------------------------------------------------------
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 93 (tp) 11 (fn)
Neg 71 (fp) 253 (tn)
--------------------------------------------------------------------------------
********************************************************************************
Report Model Metrics: xgb
Metric Value
0 Precision/PPV 0.567073
1 Average Precision 0.817957
2 Sensitivity 0.894231
3 Specificity 0.780864
4 AUC ROC 0.930051
5 Brier Score 0.165771
********************************************************************************
--------------------------------------------------------------------------------
precision recall f1-score support
0 0.96 0.78 0.86 324
1 0.57 0.89 0.69 104
accuracy 0.81 428
macro avg 0.76 0.84 0.78 428
weighted avg 0.86 0.81 0.82 428
--------------------------------------------------------------------------------
Feature names selected:
['time', 'preanti', 'str2', 'strat', 'symptom', 'treat', 'offtrt', 'cd40', 'cd420', 'cd80']
Optimal threshold used: 0.25
Important
Passing feature_selection=True in conjunction with accounting for rfe for
the pipeline_steps inside the Model` class above is necessary to print the
output of the feature names selected, thus yielding:
Feature names selected:
['time', 'preanti', 'str2', 'strat', 'symptom', 'treat', 'offtrt', 'cd40', 'cd420', 'cd80']
SHAP (SHapley Additive exPlanations)
This example demonstrates how to compute and visualize SHAP (SHapley Additive exPlanations) values for a machine learning model with a pipeline that includes feature selection. SHAP values provide insights into how individual features contribute to the predictions of a model.
Steps
The dataset is transformed through the model’s feature selection pipeline to ensure only the selected features are used for SHAP analysis.
The final model (e.g.,
XGBoostclassifier) is retrieved from the custom Model object. This is required because SHAP operates on the underlying model, not the pipeline.SHAP’s
TreeExplaineris used to explain the predictions of the XGBoost classifier.SHAP values are calculated for the transformed dataset to quantify the contribution of each feature to the predictions.
A summary plot is generated to visualize the impact of each feature across all data points.
Step 1: Transform the test data using the feature selection pipeline
## The pipeline applies preprocessing (e.g., imputation, scaling) and feature
## selection (RFE) to X_test
X_test_transformed = model_xgb.get_feature_selection_pipeline().transform(X_test)
Step 2: Retrieve the trained XGBoost classifier from the pipeline
## The last estimator in the pipeline is the XGBoost model
xgb_classifier = model_xgb.estimator[-1]
Step 3: Extract feature names from the training data, and initialize the SHAP explainer for the XGBoost classifier
## Import SHAP for model explainability
import shap
## Feature names are required for interpretability in SHAP plots
## If feature selection, Column Transformer or One Hot Encoders were used
feature_names = model.get_feature_names()
## Initialize the SHAP explainer with the model
explainer = shap.TreeExplainer(xgb_classifier)
Step 4: Compute SHAP values for the transformed test dataset and generate a summary plot of SHAP values
## Compute SHAP values for the transformed dataset
shap_values = explainer.shap_values(X_test_transformed)
Step 5: Generate a summary plot of SHAP values
## Plot SHAP values
## Summary plot of SHAP values for all features across all data points
shap.summary_plot(shap_values, X_test_transformed, feature_names=feature_names)
Feature Importance and Impact
This SHAP summary plot provides a detailed visualization of how each feature
contributes to the model’s predictions, offering insight into feature importance
and their directional effects. The X-axis represents SHAP values, which quantify
the magnitude and direction of a feature’s influence. Positive SHAP values
indicate that the feature increases the predicted output, while negative values
suggest a decrease. Along the Y-axis, features are ranked by their overall importance,
with the most influential features, such as time, positioned at the top.
Each point on the plot corresponds to an individual observation, where the color gradient reflects the feature value. Blue points represent lower feature values, while pink points indicate higher values, allowing us to observe how varying feature values affect the prediction. For example, the time feature shows a wide range of SHAP values, with higher values (pink) strongly increasing the prediction and lower values (blue) reducing it, demonstrating its critical role in driving the model’s output.
In contrast, features like hemo and age exhibit SHAP values closer to zero,
signifying a lower overall impact on predictions. Features such as homo, karnof,
and trt show more variability in their influence, indicating that their effect is
context-dependent and can significantly shift predictions in certain cases. This
plot provides a holistic view of feature behavior, enabling a deeper understanding
of the model’s decision-making process.
Multi-Class Classification
Multi-class classification involves training a model to predict one of three or more distinct classes for each instance in a dataset. Unlike binary classification, where the model predicts between two classes (e.g., positive/negative), multi-class classification applies to problems where multiple outcomes exist, such as predicting the species of flowers in the Iris dataset.
This section demonstrates how to perform multi-class classification using the
model_tuner library, with XGBoostClassifier as the base estimator
and the Iris dataset as the example.
Iris Dataset with XGBoost
The Iris dataset is a benchmark dataset commonly used for multi-class classification. It contains 150 samples from three species of Iris flowers (Setosa, Versicolour, and Virginica), with four features: sepal length, sepal width, petal length, and petal width.
Step 1: Import Necessary Libraries
import pandas as pd
import numpy as np
from model_tuner.model_tuner_utils import Model, report_model_metrics
from sklearn.impute import SimpleImputer
from xgboost import XGBClassifier
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.compose import ColumnTransformer
from sklearn.datasets import load_iris
Step 2: Load the dataset. Define X, y
data = load_iris()
X = data.data
y = data.target
X = pd.DataFrame(X)
y = pd.DataFrame(y)
Step 3: Define the preprocessing steps
Preprocessing is a crucial step in machine learning workflows to ensure the input data is properly formatted and cleaned for the model. In this case, we define a preprocessing pipeline to handle scaling and missing values in numerical features. This ensures that the data is standardized and ready for training without introducing bias from inconsistent feature ranges or missing values.
The preprocessing pipeline consists of the following components:
Numerical Transformer: A pipeline that applies:
StandardScalerfor standardizing numerical features.SimpleImputerfor imputing missing values with the mean strategy.
Column Transformer: Applies the numerical transformer to all columns and passes any remaining features through without transformation.
scalercols = X.columns
numerical_transformer = Pipeline(
steps=[
("scaler", StandardScaler()),
("imputer", SimpleImputer(strategy="mean")),
]
)
# Create the ColumnTransformer with passthrough
preprocessor = ColumnTransformer(
transformers=[
("num", numerical_transformer, scalercols),
],
remainder="passthrough",
)
Step 4: Define the estimator and hyperparameters
In this step, we configure the XGBoostClassifier as the model estimator and define its hyperparameters for multi-class classification.
We use
XGBClassifierwith theobjective="multi:softprob"parameter, which specifies multi-class classification using the softmax probability output.Assign a name to the estimator for identification in the pipeline (e.g.,
xgb_mcfor “XGBoost Multi-Class”).Enable early stopping (
early_stopping_rounds=20) to prevent overfitting by halting training if validation performance does not improve after 20 rounds.Define a hyperparameter grid for tuning:
max_depth: The maximum depth of a tree (e.g., 3, 10, 15).n_estimators: The number of boosting rounds (e.g., 5, 10, 15, 20).eval_metric: Evaluation metric (mloglossfor multi-class log-loss).verbose: Controls verbosity of output during training (1 = show progress).early_stopping_rounds: Number of rounds for early stopping.
Additional Configuration:
Disable cross-validation (
kfold=False) and calibration (calibrate=False).
estimator = XGBClassifier(objective="multi:softprob")
estimator_name = "xgb_mc"
xgbearly = True
tuned_parameters = {
f"{estimator_name}__max_depth": [3, 10, 15],
f"{estimator_name}__n_estimators": [5, 10, 15, 20],
f"{estimator_name}__eval_metric": ["mlogloss"],
f"{estimator_name}__verbose": [0],
f"{estimator_name}__early_stopping_rounds": [20],
}
kfold = False
calibrate = False
Step 5: Initialize and configure the model
After defining the preprocessing steps and estimator, the next step is to initialize the Model class from the model_tuner library. This class brings together all essential components, including the preprocessing pipeline, estimator, hyperparameters, and scoring metrics, to streamline the model training and evaluation process.
The updated configuration includes:
Name and Type:
Specify a descriptive
namefor the model (e.g., “XGB Multi Class”).Set the
model_typeto"classification"for multi-class classification.
Incorporate the
preprocessordefined earlier using theColumnTransformer, which handles scaling and imputation for numerical features.Estimator and Hyperparameters:
Link the
estimator_nameto the hyperparameter grid defined earlier (tuned_parameters).Pass the
XGBClassifieras theestimator.
Early Stopping and Cross-Validation:
Enable early stopping with
boost_early=True.Disable cross-validation with
kfold=False.
Additional Configurations:
Use
stratify_y=Truefor stratified splits.Set
multi_label=Trueto enable multi-class classification.Use
roc_auc_ovr(One-vs-Rest ROC AUC) as the scoring metric.Specify the class labels for the Iris dataset (
["1", "2", "3"]).
model_xgb = Model(
name="XGB Multi Class",
model_type="classification",
estimator_name=estimator_name,
pipeline_steps=[("ColumnTransformer", preprocessor)],
calibrate=calibrate,
estimator=estimator,
kfold=kfold,
stratify_y=True,
boost_early=xgbearly,
grid=tuned_parameters,
multi_label=True,
randomized_grid=False,
n_iter=4,
scoring=["roc_auc_ovr"],
n_jobs=-2,
random_state=42,
class_labels=["1", "2", "3"],
)
Step 6: Perform grid search parameter tuning
With the model configured, the next step is to perform grid search parameter tuning
to find the optimal hyperparameters for the XGBClassifier. The
grid_search_param_tuning method will iterate over all combinations of
hyperparameters specified in tuned_parameters, evaluate each one using the
specified scoring metric, and select the best performing set.
This method will:
Split the Data: The data will be split into training and validation sets. Since
stratify_y=True, the class distribution will be maintained across splits.Iterate Over Hyperparameters: All combinations of hyperparameters defined in
tuned_parameterswill be tried sincerandomized_grid=False.Early Stopping: With
boost_early=Trueandearly_stopping_roundsset in the hyperparameters, the model will stop training early if the validation score does not improve.Scoring: The model uses
roc_auc_ovr(One-vs-Rest ROC AUC) as the scoring metric suitable for multi-class classification.Select Best Model: The hyperparameter set that yields the best validation score will be selected.
To execute the grid search, simply call:
model.grid_search_param_tuning(X, y)
Pipeline Steps:
┌───────────────────────────────────────────────────────────┐
│ Step 1: preprocess_column_transformer_ColumnTransformer │
│ ColumnTransformer │
└───────────────────────────────────────────────────────────┘
│
▼
┌───────────────────────────────────────────────────────────┐
│ Step 2: xgb_mc │
│ XGBClassifier │
└───────────────────────────────────────────────────────────┘
100%|██████████| 12/12 [00:00<00:00, 22.10it/s]Best score/param set found on validation set:
{'params': {'xgb_mc__early_stopping_rounds': 20,
'xgb_mc__eval_metric': 'mlogloss',
'xgb_mc__max_depth': 10,
'xgb_mc__n_estimators': 10},
'score': 0.9666666666666668}
Best roc_auc_ovr: 0.967
Step 7: Generate data splits
Once the best hyperparameters are identified through grid search, the next step
is to generate the training, validation, and test splits. The Model class
provides built-in methods for creating these splits while maintaining the class
distribution (as specified by stratify_y=True).
Use the following code to generate the splits:
## Get the training, validation, and test data
X_train, y_train = model_xgb.get_train_data(X, y)
X_valid, y_valid = model_xgb.get_valid_data(X, y)
X_test, y_test = model_xgb.get_test_data(X, y)
Description of Splits:
Training Data (
X_train,y_train): Used to train the model.Validation Data (
X_valid,y_valid): Used during training for monitoring and fine-tuning, including techniques like early stopping.Test Data (
X_test,y_test): Reserved for evaluating the final performance of the trained model.
These splits ensure that each phase of model development (training, validation, and testing) is performed on separate portions of the dataset, providing a robust evaluation pipeline.
Step 8: Fit the model
After generating the data splits, the next step is to train the model using the
training data and validate its performance on the validation data during training.
The fit method in the Model class handles this process seamlessly,
leveraging the best hyperparameters found during grid search.
Use the following code to fit the model:
model_xgb.fit(
X_train,
y_train,
validation_data=[X_valid, y_valid],
)
Note
Training Data (
X_train,y_train): The model is trained on this data to learn patterns.Validation Data (
X_valid,y_valid): During training, the model monitors its performance on this data to avoid overfitting and apply techniques like early stopping.Early Stopping: If
boost_early=Trueandearly_stopping_roundsis defined, training will halt early when validation performance stops improving.
This step ensures that the model is fitted using the best configuration from the grid search and optimized for generalization. With the model trained, proceed to Step 9 to evaluate its performance on validation and test datasets.
Step 9: Return metrics (optional)
Once the model is trained, you can evaluate its performance on the validation
and test datasets by returning key metrics. The return_metrics method from
the Model class calculates and displays metrics like ROC AUC, precision, recall, and F1-score.
Use the following code to return metrics:
# Evaluate on validation data
print("Validation Metrics")
model_xgb.return_metrics(
X_valid,
y_valid,
optimal_threshold=True,
)
# Predict probabilities for the test data
y_prob = model.predict_proba(X_test)
# Evaluate on test data
print("Test Metrics")
model_xgb.return_metrics(
X_test,
y_test,
optimal_threshold=True,
)
Validation Metrics
--------------------------------------------------------------------------------
1
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 19 (tn) 1 (fp)
Neg 0 (fn) 10 (tp)
--------------------------------------------------------------------------------
2
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 19 (tn) 1 (fp)
Neg 2 (fn) 8 (tp)
--------------------------------------------------------------------------------
3
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 19 (tn) 1 (fp)
Neg 1 (fn) 9 (tp)
--------------------------------------------------------------------------------
precision recall f1-score support
0 0.91 1.00 0.95 10
1 0.89 0.80 0.84 10
2 0.90 0.90 0.90 10
accuracy 0.90 30
macro avg 0.90 0.90 0.90 30
weighted avg 0.90 0.90 0.90 30
--------------------------------------------------------------------------------
Test Metrics
--------------------------------------------------------------------------------
1
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 18 (tn) 2 (fp)
Neg 0 (fn) 10 (tp)
--------------------------------------------------------------------------------
2
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 19 (tn) 1 (fp)
Neg 2 (fn) 8 (tp)
--------------------------------------------------------------------------------
3
Predicted:
Pos Neg
--------------------------------------------------------------------------------
Actual: Pos 20 (tn) 0 (fp)
Neg 1 (fn) 9 (tp)
--------------------------------------------------------------------------------
precision recall f1-score support
0 0.83 1.00 0.91 10
1 0.89 0.80 0.84 10
2 1.00 0.90 0.95 10
accuracy 0.90 30
macro avg 0.91 0.90 0.90 30
weighted avg 0.91 0.90 0.90 30
--------------------------------------------------------------------------------
{'Classification Report': {'0': {'precision': 0.8333333333333334,
'recall': 1.0,
'f1-score': 0.9090909090909091,
'support': 10.0},
'1': {'precision': 0.8888888888888888,
'recall': 0.8,
'f1-score': 0.8421052631578948,
'support': 10.0},
'2': {'precision': 1.0,
'recall': 0.9,
'f1-score': 0.9473684210526316,
'support': 10.0},
'accuracy': 0.9,
'macro avg': {'precision': 0.9074074074074074,
'recall': 0.9,
'f1-score': 0.8995215311004786,
'support': 30.0},
'weighted avg': {'precision': 0.9074074074074073,
'recall': 0.9,
'f1-score': 0.8995215311004785,
'support': 30.0}},
'Confusion Matrix': array([[[18, 2],
[ 0, 10]],
[[19, 1],
[ 2, 8]],
[[20, 0],
[ 1, 9]]])}
Report Model Metrics
You can summarize and display the model’s performance metrics using the
report_model_metrics function. This function computes key metrics like
precision, recall, F1-score, and ROC AUC for each class, as well as macro and weighted averages.
Use the following code:
metrics_df = report_model_metrics(
model=model_xgb,
X_valid=X_test,
y_valid=y_test,
threshold=next(iter(model_xgb.threshold.values())),
)
print(metrics_df)
0 Precision/PPV 0.833333
0 Sensitivity/Recall 1.000000
0 F1-Score 0.909091
1 Precision/PPV 0.888889
1 Sensitivity/Recall 0.800000
1 F1-Score 0.842105
2 Precision/PPV 1.000000
2 Sensitivity/Recall 0.900000
2 F1-Score 0.947368
macro avg Precision/PPV 0.907407
macro avg Sensitivity/Recall 0.900000
macro avg F1-Score 0.899522
weighted avg Precision/PPV 0.907407
weighted avg Sensitivity/Recall 0.900000
weighted avg F1-Score 0.899522
Weighted Average Precision 0.907407
Weighted Average Recall 0.900000
Multiclass AUC ROC 0.933333
Note
Validation Metrics: Provide insights into how well the model performed during training and tuning on unseen validation data.
Test Metrics: Assess the final model’s generalization performance on completely unseen test data.
predict_proba: Outputs the predicted probabilities for each class, useful for calculating metrics like ROC AUC or understanding the model’s confidence in its predictions.
By examining these metrics, you can evaluate the model’s strengths and weaknesses and determine if further fine-tuning or adjustments are necessary.
Step 10: Predict probabilities and generate predictions
As an additional step, you can use the trained model to predict probabilities and generate predictions for the test data. This is particularly useful for analyzing model outputs or evaluating predictions with custom thresholds.
Use the following code:
## Predict probabilities for the test data
y_prob = model.predict_proba(X_test)[:, 1]
## Predict class labels using the optimal threshold
y_pred = model.predict(X_test, optimal_threshold=True)
# Print results
print(f"Predicted Probabilities: \n {y_prob}")
print()
print(f"Predictions: \n {y_pred}")
Predicted Probabilities:
[0.961671 0.02298635 0.749543 0.02298635 0.0244073 0.02298635
0.94500786 0.02298635 0.0227305 0.02298635 0.14078036 0.32687086
0.94500786 0.961671 0.95576227 0.02298635 0.02298635 0.02298635
0.961671 0.0244073 0.0227305 0.02298635 0.02298635 0.38560066
0.02298635 0.02298635 0.961671 0.0227305 0.0227305 0.4547262 ]
Predictions:
[1 0 1 0 0 0 1 0 0 0 0 0 1 1 1 0 0 0 1 0 0 0 0 0 0 0 1 0 0 0]
Note
Predicted Probabilities (
predict_proba): Returns the probabilities for each class. The[:, 1]selects the probabilities for the second class (or one of interest).Predicted Labels (
predict): Generates class predictions using the optimal threshold, which is tuned during grid search or based on the scoring metric.Optimal Threshold: When
optimal_threshold=True, the model uses the threshold that maximizes a selected performance metric (e.g., F1-score or ROC AUC) instead of the default threshold of 0.5.Analysis: Inspecting probabilities and predictions helps to interpret the model’s confidence and accuracy in making decisions.
This step allows for a deeper understanding of the model’s predictions and can be used to fine-tune decision thresholds or evaluate specific cases.
Regression
Here is an example of using the Model class for a regression task with XGBoost on the California Housing dataset.
The California Housing dataset, available in the sklearn library, is a commonly used benchmark dataset for regression problems. It contains features such as median income, housing age, and population, which are used to predict the median house value for California districts.
In this example, we leverage the Model class to:
Set up an XGBoost regressor as the estimator.
Define a hyperparameter grid for tuning the model.
Preprocess the dataset, train the model, and evaluate its performance using the \(R^2\) metric.
The workflow highlights how the Model class simplifies regression tasks, including hyperparameter tuning, and performance evaluation.
California Housing with XGBoost
Step 1: Import necessary libraries
import pandas as pd
import numpy as np
from xgboost import XGBRegressor
from sklearn.impute import SimpleImputer
from sklearn.datasets import fetch_california_housing
from model_tuner import Model
Step 2: Load the dataset
## Load the California Housing dataset
data = fetch_california_housing()
X = pd.DataFrame(data.data, columns=data.feature_names)
y = pd.Series(data.target, name="target")
Step 3: Create an instance of the XGBRegressor
xgb_name = "xgb"
xgb = XGBRegressor(random_state=222)
Step 4: Define Hyperparameters for XGBRegressor
In this step, we configure the XGBRegressor for a regression task.
The hyperparameter grid includes key settings to control the learning process,
tree construction, and generalization performance of the model.
The hyperparameter grid and model configuration are defined as follows:
tuned_parameters_xgb = [
{
f"{xgb_name}__learning_rate": [0.1, 0.01, 0.05],
f"{xgb_name}__n_estimators": [100, 200, 300],
f"{xgb_name}__max_depth": [3, 5, 7][:1],
f"{xgb_name}__subsample": [0.8, 1.0][:1],
f"{xgb_name}__colsample_bytree": [0.8, 1.0][:1],
f"{xgb_name}__eval_metric": ["logloss"],
f"{xgb_name}__early_stopping_rounds": [10],
f"{xgb_name}__tree_method": ["hist"],
f"{xgb_name}__verbose": [0],
}
]
xgb_definition = {
"clc": xgb,
"estimator_name": xgb_name,
"tuned_parameters": tuned_parameters_xgb,
"randomized_grid": False,
"early": True,
}
model_definition = {xgb_name: xgb_definition}
Key Configurations
learning_rate: Controls the contribution of each boosting round to the final prediction.n_estimators: Specifies the total number of boosting rounds (trees).max_depth: Limits the depth of each tree to prevent overfitting.subsample: Fraction of training data used for fitting each tree, introducing randomness to improve generalization.colsample_bytree: Fraction of features considered for each boosting round.eval_metric: Specifies the evaluation metric to monitor during training (e.g.,"logloss").early_stopping_rounds: Stops training if validation performance does not improve for a set number of rounds.tree_method: Chooses the algorithm used for tree construction ("hist"for histogram-based methods, optimized for speed).verbose: Controls output display during training (set to0for silent mode).
Step 5: Initialize and configure the Model
XGBRegressor inherently handles missing values (NaN) without requiring explicit
imputation strategies. During training, XGBoost treats missing values as a
separate category and learns how to route them within its decision trees.
Therefore, passing a SimpleImputer or using an imputation strategy is unnecessary
when using XGBRegressor.
kfold = False
calibrate = False
## Define model object
model_type = "xgb"
clc = model_definition[model_type]["clc"]
estimator_name = model_definition[model_type]["estimator_name"]
## Set the parameters by cross-validation
tuned_parameters = model_definition[model_type]["tuned_parameters"]
rand_grid = model_definition[model_type]["randomized_grid"]
early_stop = model_definition[model_type]["early"]
model_xgb = Model(
name=f"xgb_{model_type}",
estimator_name=estimator_name,
model_type="regression",
calibrate=calibrate,
estimator=clc,
kfold=kfold,
stratify_y=False,
grid=tuned_parameters,
randomized_grid=rand_grid,
boost_early=early_stop,
scoring=["r2"],
random_state=222,
n_jobs=2,
)
Step 6: Perform grid search parameter tuning and retrieve split data
To execute the grid search, simply call:
model_xgb.grid_search_param_tuning(X, y)
## Get the training, validation, and test data
X_train, y_train = model_xgb.get_train_data(X, y)
X_valid, y_valid = model_xgb.get_valid_data(X, y)
X_test, y_test = model_xgb.get_test_data(X, y)
With the model configured, the next step is to perform grid search parameter tuning
to find the optimal hyperparameters for the XGBRegressor. The
grid_search_param_tuning method will iterate over all combinations of
hyperparameters specified in tuned_parameters_xgb, evaluate each one using the
specified scoring metric, and select the best performing set.
This method will:
Split the Data: The data will be split into training and validation sets. Since
stratify_y=False, the class distribution will not be maintained across splits.Iterate Over Hyperparameters: All combinations of hyperparameters defined in
tuned_parameters_xgbwill be tried sincerandomized_grid=False.Early Stopping: With
boost_early=Trueandearly_stopping_roundsset in the hyperparameters, the model will stop training early if the validation score does not improve.Scoring: The model uses \(R^2\) as the scoring metric, which is suitable for evaluating regression models.
Select Best Model: The hyperparameter set that yields the best validation score based on the specified metric (\(R^2\)) will be selected.
Pipeline Steps:
┌────────────────┐
│ Step 1: xgb │
│ XGBRegressor │
└────────────────┘
100%|██████████| 9/9 [00:22<00:00, 2.45s/it]Best score/param set found on validation set:
{'params': {'xgb__colsample_bytree': 0.8,
'xgb__early_stopping_rounds': 10,
'xgb__eval_metric': 'logloss',
'xgb__learning_rate': 0.1,
'xgb__max_depth': 3,
'xgb__n_estimators': 67,
'xgb__subsample': 0.8,
'xgb__tree_method': 'hist'},
'score': 0.7651490279157868}
Best r2: 0.765
Step 7: Fit the model
model_xgb.fit(
X_train,
y_train,
validation_data=[X_valid, y_valid],
)
Step 8: Return metrics (optional)
Validation Metrics
********************************************************************************
{'Explained Variance': 0.7647451659057567,
'Mean Absolute Error': 0.3830825326824073,
'Mean Squared Error': 0.3066172248224347,
'Median Absolute Error': 0.2672762813568116,
'R2': 0.7647433075624044,
'RMSE': 0.5537302816556403}
********************************************************************************
Test Metrics
********************************************************************************
{'Explained Variance': 0.7888942913974833,
'Mean Absolute Error': 0.3743548199982513,
'Mean Squared Error': 0.28411432705731066,
'Median Absolute Error': 0.26315186452865597,
'R2': 0.7888925135381788,
'RMSE': 0.533023758436067}
********************************************************************************
{'Explained Variance': 0.7888942913974833,
'R2': 0.7888925135381788,
'Mean Absolute Error': 0.3743548199982513,
'Median Absolute Error': 0.26315186452865597,
'Mean Squared Error': 0.28411432705731066,
'RMSE': 0.533023758436067}
Performance Evaluation Metrics
Using report_model_metrics()
The report_model_metrics() method provides detailed insights into model performance, including metrics such as precision, recall, sensitivity, specificity, and AUC-ROC. For regression models, it includes key metrics such as Mean Absolute Error (MAE), Mean Squared Error (MSE), Root Mean Squared Error (RMSE), R² Score, and Explained Variance.
While this method is integrated into return_metrics(), it can also be invoked independently for
custom evaluation workflows. For example, it can be used to focus on specific metrics or to analyze
a subset of the data.
Using return_metrics()
A key feature of return_metrics() is its ability to retrieve and print the
threshold value used to train the model. This threshold can be passed directly
into report_model_metrics() for consistent evaluation.
Threshold Tuning
Model thresholding is a critical concept in classification tasks, allowing you to fine-tune the decision boundary for predicting positive or negative classes. Instead of relying on the default threshold of 0.5, which may not suit all datasets or evaluation metrics, thresholds can be adjusted to optimize metrics like precision, recall, or F1-score based on your specific objectives.
The model.threshold attribute provides a dictionary where each scoring metric
is paired with its corresponding optimal threshold, enabling precise control over
predictions. This is particularly useful in applications where the cost of false
positives and false negatives differs significantly.
For example:
## Accessing the optimal thresholds for each scoring metric
print(model_xgb.threshold)
{'roc_auc': 0.25}
When to Use Custom Thresholds:
Imbalanced Datasets: Adjusting thresholds can help mitigate the effects of class imbalance by prioritizing recall or precision for the minority class.
Domain-Specific Goals: In medical diagnostics, for instance, you might prefer a lower threshold to maximize sensitivity (recall) and minimize false negatives.
Optimizing for Specific Metrics: If your primary evaluation metric is F-beta, tuning the threshold ensures better alignment with your goals.
How to Automatically Tune Thresholds
The optimal threshold can be automatically tuned by enabling F-beta optimization
during parameter tuning. This can be done by setting f1_beta_tune=True in
the grid_search_param_tuning() method:
# Automatically tune thresholds for F-beta optimization
model_xgb.grid_search_param_tuning(X, y, f1_beta_tune=True)
After tuning, the optimal thresholds will be stored in the model.threshold
attribute for each scoring metric:
## Retrieve the optimal threshold for a specific metric
threshold = model_xgb.threshold['roc_auc']
Using threshold in report_model_metrics()
After calling return_metrics() with optimal_threshold=True,
you can reuse the threshold in report_model_metrics() as shown below:
threshold = model_xgb.threshold['roc_auc'] # Retrieve the optimal threshold
model_xgb.report_model_metrics(X=X_valid, y=y_valid, threshold=threshold)
Reporting Threshold in return_metrics
The return_metrics method provides the flexibility to retrieve and print the
threshold used during model evaluation, enabling seamless reuse in other methods
or manual experimentation. When print_threshold=True is specified, the
threshold will be included as part of the output, making it easy to reference
and apply in subsequent analyses.
Example:
# Retrieve metrics and threshold using return_metrics
model_xgb.return_metrics(
X=X_valid,
y=y_valid,
optimal_threshold=True,
print_threshold=True,
model_metrics=True
)
By including print_threshold=True, the optimal threshold used for predictions
is displayed, ensuring transparency and providing a valuable reference for further
evaluations or custom workflows.
Classification report (optional)
A call to print(model_xgb.classification_report) will
output the classification report as follows:
print(model_xgb.classification_report)
precision recall f1-score support
0 0.91 0.94 0.92 324
1 0.79 0.71 0.75 104
accuracy 0.88 428
macro avg 0.85 0.82 0.84 428
weighted avg 0.88 0.88 0.88 428
Bootstrap Metrics
The bootstrapper.py module provides utility functions for input type checking,
data resampling, and evaluating bootstrap metrics.
- check_input_type(x)
Validates and normalizes the input type for data processing. Converts NumPy arrays, Pandas Series, and DataFrames into a standard Pandas DataFrame with a reset index.
- Parameters:
x (array-like) – Input data (NumPy array, Pandas Series, or DataFrame).
- Returns:
Normalized input as a Pandas DataFrame.
- Return type:
pandas.DataFrame
- Raises:
ValueError – If the input type is not supported.
- sampling_method(y, n_samples, stratify=False, balance=False, class_proportions=None)
Resamples a dataset based on specified options for balancing, stratification, or custom class proportions.
- Parameters:
y (pandas.Series) – Target variable to resample.
n_samples (int) – Number of samples to draw.
stratify (bool, optional) – Whether to stratify based on the provided target variable.
balance (bool, optional) – Whether to balance class distributions equally.
class_proportions (dict, optional) – Custom proportions for each class. Must sum to 1.
- Returns:
Resampled target variable.
- Return type:
pandas.DataFrame
- Raises:
ValueError – If class proportions do not sum to 1.
- evaluate_bootstrap_metrics(model=None, X=None, y=None, y_pred_prob=None, n_samples=500, num_resamples=1000, metrics=['roc_auc', 'f1_weighted', 'average_precision'], random_state=42, threshold=0.5, model_type='classification', stratify=None, balance=False, class_proportions=None)
Evaluates classification or regression metrics on bootstrap samples using a pre-trained model or pre-computed predictions.
- Parameters:
model (object, optional) – Pre-trained model with a
predict_probamethod. Required ify_pred_probis not provided.X (array-like, optional) – Input features. Not required if
y_pred_probis provided.y (array-like) – Ground truth labels.
y_pred_prob (array-like, optional) – Pre-computed predicted probabilities.
n_samples (int, optional) – Number of samples per bootstrap iteration. Default is 500.
num_resamples (int, optional) – Number of bootstrap iterations. Default is 1000.
metrics (list of str) – List of metrics to calculate (e.g.,
"roc_auc","f1_weighted").random_state (int, optional) – Random seed for reproducibility. Default is 42.
threshold (float, optional) – Classification threshold for probability predictions. Default is 0.5.
model_type (str) – Specifies the task type, either
"classification"or"regression".stratify (pandas.Series, optional) – Variable for stratified sampling.
balance (bool, optional) – Whether to balance class distributions.
class_proportions (dict, optional) – Custom class proportions for sampling.
- Returns:
DataFrame with mean and confidence intervals for each metric.
- Return type:
pandas.DataFrame
- Raises:
ValueError – If invalid parameters or metrics are provided.
RuntimeError – If sample size is insufficient for metric calculation.
Note
The model_tuner_utils.py module includes utility functions for evaluating bootstrap metrics in the context of model tuning.
- return_bootstrap_metrics(X_test, y_test, metrics, threshold=0.5, num_resamples=500, n_samples=500, balance=False)
Evaluates bootstrap metrics for a trained model using the test dataset. This function supports both classification and regression tasks by leveraging evaluate_bootstrap_metrics to compute confidence intervals for the specified metrics.
- Parameters:
X_test (pandas.DataFrame) – Test dataset features.
y_test (pandas.Series or pandas.DataFrame) – Test dataset labels.
metrics (list of str) – List of metric names to calculate (e.g.,
"roc_auc","f1_weighted").threshold (float, optional) – Threshold for converting predicted probabilities into class predictions. Default is 0.5.
num_resamples (int, optional) – Number of bootstrap iterations. Default is 500.
n_samples (int, optional) – Number of samples per bootstrap iteration. Default is 500.
balance (bool, optional) – Whether to balance the class distribution during resampling. Default is False.
- Returns:
DataFrame containing mean and confidence intervals for the specified metrics.
- Return type:
pandas.DataFrame
- Raises:
ValueError – If
X_testory_testare not provided as Pandas DataFrames or if unsupported input types are specified.
Bootstrap metrics example
Continuing from the model output object (model_xgb) from the regression example above, we leverage the return_bootstrap_metrics method from model_tuner_utils.py to print bootstrap performance metrics (\(R^2\) and \(\text{explained variance}\)) at 95% confidence levels as shown below:
print("Bootstrap Metrics")
model_xgb.return_bootstrap_metrics(
X_test=X_test,
y_test=y_test,
metrics=["r2", "explained_variance"],
n_samples=30,
num_resamples=300,
)
Bootstrap Metrics
100%|██████████| 300/300 [00:00<00:00, 358.05it/s]
Metric Mean 95% CI Lower 95% CI Upper
0 r2 0.781523 0.770853 0.792193
1 explained_variance 0.788341 0.777898 0.798785